Real-Time Determination of the Cell-Cycle Position of Individual Cells within Live Tumors Using FUCCI Cell-Cycle Imaging
Abstract
:1. Introduction
1.1. The Development of Intravital “Orthotopic” Imaging and a Fluorescent Cell Cycle Probe
1.2. FUCCI (Fluorescence Ubiquitination Cell Cycle Indicator) Repoters
2. Longitudinal Intravital Imaging of an Orthotopic Metastatic Liver Tumor Model with FUCCI
2.1. Skin-Window System
2.2. Minimal Organ-Stabilization System Using Styrofoam and Pins
2.3. Cell-Cycle Distribution within a Tumor
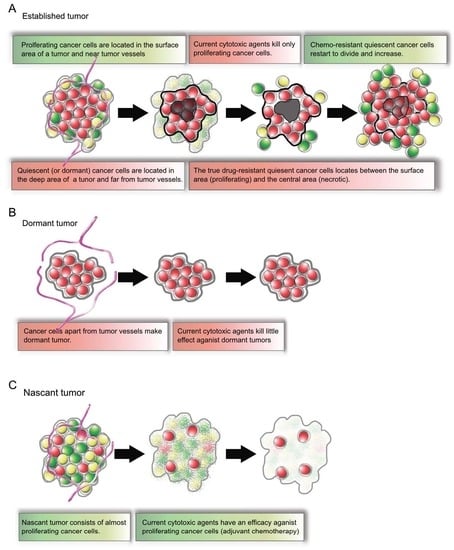
2.4. Established Tumors Consist of a Vast Majority of Quiescent Cancer Cells
3. Intravital Orthotopic FUCCI Imaging Reveals the Relationship between Cell Cycle Phase of Cancer Cells and the Juxtaposition of Tumor Blood Vessels
4. Intravital Orthotopic FUCCI Imaging Reveals that Quiescent Cancer Cells are Resistant to Conventional Chemotherapy
5. Intravital Orthotopic FUCCI Imaging Unveils the Adverse Effect of Irradiation Therapy
6. Intravital Orthotopic FUCCI Imaging Identifies Cell Cycle-Related Genes
7. New Cell-Cycle-Based Approaches to Treatment of Solid Tumor: Decoy, Trap, and Shoot Therapy
8. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Hoffman, R.M. The multiple uses of fluorescent proteins to visualize cancer in vivo. Nat. Rev. Cancer 2005, 10, 796–806. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, R.M.; Yang, M. Color-coded fluorescence imaging of tumor host interactions. Nat Protoc. 2006, 1, 928–935. [Google Scholar] [CrossRef] [PubMed]
- Newman, R.H.; Fosbrink, M.D.; Zhang, J. Genetically encodable fluorescent biosensors for tracking signaling dynamics in living cells. Chem. Rev. 2011, 111, 3614–3666. [Google Scholar] [CrossRef] [PubMed]
- Condeelis, J.; Segall, J.E. Intravital imaging of cell movement in tumours. Nat. Rev. Cancer 2003, 8, 921–930. [Google Scholar] [CrossRef] [PubMed]
- Chishima, T.; Miyagi, Y.; Wang, X.; Yamaoka, H.; Shimada, H.; Moosa, A.R.; Hoffman, R.M. Cancer invasion and micrometastasis visualized in live tissue by green fluorescent protein expression. Cancer Res. 1997, 57, 2042–2047. [Google Scholar] [PubMed]
- Chishima, T.; Miyagi, Y.; Wang, X.; Yamaoka, H.; Shimada, H.; Moosa, A.R.; Hoffman, R.M. Visualization of the metastatic process by green fluorescent protein expression. Anticancer Res. 1997, 17, 2377–2384. [Google Scholar] [PubMed]
- Chishima, T.; Miyagi, Y.; Wang, X.; Yamaoka, H.; Shimada, H.; Moosa, A.R.; Hoffman, R.M. Metastatic patterns of lung cancer visualized live and in process by green fluorescence protein expression. Clin. Exp. Metastasis 1997, 15, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Hasegawa, S.; Jiang, P.; Wang, X.; Tan, Y.; Chishima, T.; Shimada, H.; Moosa, A.R.; Hoffman, R.M. Widespread skeletal metastatic potential of human lung cancer revealed by green fluorescent protein expression. Cancer Res. 1998, 58, 4217–4221. [Google Scholar] [PubMed]
- Yang, M.; Jiang, P.; Sun, F.X.; Hasegawa, S.; Baranov, E.; Chishima, T.; Shimada, H.; Moosa, A.R.; Hoffman, R.M. A fluorescent orthotopic bone metastasis model of human prostate cancer. Cancer Res. 1999, 59, 781–786. [Google Scholar] [PubMed]
- Yang, M.; Jiang, P.; An, Z.; Baranov, E.; Li, L.; Hasegawa, S.; Chishima, T.; Shimada, H.; Moosa, A.R.; Hoffman, R.M. Genetically fluorescent melanoma bone and organ metastasis models. Clin. Cancer Res. 1999, 5, 3549–3559. [Google Scholar] [PubMed]
- Yang, M.; Chishima, T.; Wang, X.; Baranov, E.; Shimada, H.; Moosa, A.R.; Hoffman, R.M. Multi-organ metastatic capability of Chinese hamster ovary cells revealed by green fluorescent protein (GFP) expression. Clin. Exp. Metastasis 1999, 17, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Bouvet, M.; Tsuji, K.; Yang, M.; Jiang, P.; Moosa, A.R.; Hoffman, R.M. In vivo color-coded imaging of the interaction of colon cancer cells and splenocytes in the formation of liver metastasis. Cancer Res. 2006, 66, 11293–11297. [Google Scholar] [CrossRef] [PubMed]
- Naumov, G.N.; Wilson, S.M.; MacDonald, I.C.; Schmidt, E.E.; Morris, V.L.; Groom, A.C.; Hoffman, R.M.; Chambers, A.F. Cellular expression of green fluorescent protein, coupled with high-resolution in vivo videomicroscopy, to monitor steps in tumor metastasis. J. Cell Sci. 1999, 112, 1835–1842. [Google Scholar] [PubMed]
- Yang, M.; Baranov, E.; Moosa, A.R.; Penman, S.; Hoffman, R.M. Visualizing gene expression by whole-body fluorescence imaging. Proc. Natl. Acad. Sci. USA 2000, 97, 12278–12282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, M.; Baranov, E.; Jiang, P.; Sun, F.X.; Li, X.M.; Li, L.; Hasegawa, S.; Chishima, T.; Shimada, H.; Moosa, A.R.; et al. Whole-body optical imaging of green fluorescent protein-expressing tumors and metastasis. Proc. Natl. Acad. Sci. USA 2000, 97, 1206–1211. [Google Scholar] [CrossRef] [PubMed]
- Tsien, R. The green fluorescence protein. Anuu. Rev. Biochem. 1998, 67, 509–554. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, K.; Yang, M.; Jiang, P.; Yamamoto, N.; Xu, M.; Amoh, Y.; Tsuji, K.; Bouvet, M.; Tsuchiya, H.; Tomita, K.; et al. Development of real-time subcellular dynamics multicolor imaging of cancer-cell tracking in live mice with a variable-magnification whole-mouse imaging system. Cancer Res. 2006, 66, 4208–4214. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Baranov, E.; Li, X.M.; Wang, J.W.; Jiang, P.; Li, L.; Moosa, A.R.; Penman, S.; Hoffman, R.M. Whole-body and intravital optical imaging of angiogenesis in orthotopiccally implanted tumors. Proc. Natl. Acad. Sci. USA 2001, 98, 2616–2621. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Baranov, E.; Wang, J.W.; Jiang, P.; Wang, X.; Sun, F.X.; Bouvet, M.; Moosa, A.R.; Penman, S.; Hoffman, R.M. Direct external imaging of nascent cancer, tumor progression, angiogenesis, and metastasis on internal organs in the fluorescent orthotopic model. Proc. Natl. Acad. Sci. USA 2002, 99, 3824–3829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouvet, M.; Wang, J.; Nardin, S.R.; Nassirpour, R.; Yang, M.; Baranov, E.; Jiang, P.; Moosa, A.R.; Hoffman, R.M. Real-time optical imaging of primary tumor. Cancer Res. 2002, 62, 1534–1540. [Google Scholar] [PubMed]
- Yamamoto, N.; Jiang, P.; Yang, M.; Xu, M.; Yamauchi, K.; Tsuchiya, H.; Tomita, K.; Geoffrey, M.; Abdool, R.W.; Moosa, A.R.; et al. Cellular dynamics visualized in live cells in vitro and in vivo by differential dual-color nuclear-cytoplasmic fluorescent-protein expression. Cancer Res. 2004, 64, 4251–4256. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Jiang, P.; Hoffman, R.M. Whole-body subcellular multicolor imaging of tumor-host interaction and drug response in real time. Cancer Res. 2007, 67, 5195–5200. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, N.; Yang, M.; Jiang, P.; Xu, M.; Tsuchiya, H.; Tomita, K.; Moosa, A.R.; Hoffman, R.M. Determination of clonality of metastasis by cell-specific color-coded fluorescent-protein imaging. Cancer Res. 2003, 63, 7785–7790. [Google Scholar] [PubMed]
- Yang, M.; Reynoso, J.; Jiang, P.; Li, L.; Moosa, A.R.; Hoffman, R.M. Transgenic nude mouse with ubiquitous green fluorescent protein expression as a host for human tumors. Cancer Res. 2004, 64, 8651–8656. [Google Scholar] [CrossRef] [PubMed]
- Amoh, Y.; Yang, M.; Li, L.; Reynoso, J.; Bouvet, M.; Moossa, A.R.; Katsuoka, K.; Hoffman, R.M. Nestin-linked green fluorescent protein transgenic nude mouse for imaging human tumor angiogenesis. Cancer Res. 2005, 65, 5352–5357. [Google Scholar] [CrossRef] [PubMed]
- Schepers, A.G.; Snippert, H.J.; Stange, D.E.; van den Born, M.; van Es, J.H.; van de Wetering, M.; Clevers, H. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science 2012, 337, 730–735. [Google Scholar] [CrossRef] [PubMed]
- Livet, J.; Weissman, A.; Kang, H.; Draft, W.; Lu, J.; Bennis, R.A.; Sanes, J.R.; Lichtman, J.W. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 2007, 450, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Timpson, P.; McGhee, E.J.; Anderson, K. Imaging molecular dynamics in vivo-from cell biology to animal models. J. Cell Sci. 2011, 124, 2877–2890. [Google Scholar] [CrossRef] [PubMed]
- Nobis, M.; Warren, S.C.; Luca, M.C.; Murphy, K.J.; Herrrman, D.; Timpson, P. Molecular mobility and activity in an intravital imaging setting-implications for cancer progression and targeting. J. Cell Sci. 2018, in press. [Google Scholar] [CrossRef] [PubMed]
- Weissleder, R.; Pittet, M.J. Imaging in the era of molecular oncology. Nature 2008, 452, 580–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carragher, N. Live cell in vitro and in vivo imaging applications: Accelerating drug discovery. Pharmaceutics 2011, 3, 141–170. [Google Scholar]
- Kamb, A. What’s wrong with our cancer models? Nat. Rev. Drug Discov. 2005, 4, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Beerling, E.; Ritsma, L.; Vrisekoop, N.; Derksen, P.W.; van Rheenen, J. Intravital microscopy: New insights into metastasis of tumors. J. Cell Sci. 2011, 124, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Barretto, R.P.; Ko, T.H.; Jung, J.C.; Wang, T.J.; Capps, G.; Waters, A.C.; Ziv, Y.; Attardo, A.; Recht, L.; Schnitzer, M.J. Time-lapse imaging of disease progression in deep brain areas using fluorescence microendoscopy. Nat. Med. 2011, 17, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Kedrin, D. Intravital imaging of metastatic behavior through a mammary imaging window. Nat. Methods 2008, 5, 1019–1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakaue-Sawano, A.; Kurokawa, H.; Morimura, T.; Tanyu, A.; Osawa, H.; Kashiwagi, S.; Fukami, K.; Miyata, T.; Miyoshi, H.; Imamura, T.; et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 2008, 132, 487–489. [Google Scholar] [CrossRef] [PubMed]
- Tomura, M.; Sakaue-Sawano, A.; Mori, Y.; Takase-Utsuji, M.; Hata, A.; Ohtawa, K.; Kanagawa, A.; Miyawaki, A. Contrasting quiescent G0 phase with mitotic cell cycling in the mouse immune system. PLoS ONE 2013, 8, e73801. [Google Scholar] [CrossRef] [PubMed]
- Sakaue-Sawano, A.; Kobayashi, T.; Ohtawa, K.; Miyawaki, A. Drug-induced cell cycle modulation leading to cell-cycle arrest, nuclear mis-segregation, or endoreplication. BMC Cell Biol. 2011, 12, 2. [Google Scholar] [CrossRef] [PubMed]
- Bajar, B.T.; Lam, A.J.; Badiee, R.K.; Oh, Y.H.; Chu, J.; Zhou, X.X.; Kim, N.; Kim, B.B.; Chung, M.; Yablonovitch, A.L.; et al. Fluorescent indicators for simultaneous reporting of all four cell cycle phases. Nat. Methods 2016, 13, 993–996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakaue-Sawano, A.; Yo, M.; Komatsu, N.; Hiratsuka, T.; Kogure, T.; Hoshida, T.; Goshima, N.; Matsuda, M.; Miyoshi, H.; Miyawaki, A. Genetically Encoded Tools for Optical Dissection of the Mammalian Cell Cycle. Mol. Cell 2017, 68, 626–640. [Google Scholar] [CrossRef] [PubMed]
- Oki, T.; Nishimura, K.; Kitaura, J.; Togami, K.; Maehara, A.; Izawa, K. A novel cell-cycle-indicator, mVenus-p27K-, identifies quiescent cells and visualizes G0-G1 transition. Sci. Rep. 2014, 4, 4012. [Google Scholar] [CrossRef] [PubMed]
- Chittajallu, D.R.; Florian, S.; Kohler, R.; Iwamoto, Y.; Orth, J.D.; Weissleder, R.; Danuse, G.; Mitchison, T.J. In vivo cell-cycle profiling in xenograft tumors by quantitative intravital microscopy. Nat. Methods 2015, 12, 577–585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritsma, L.; Steller, E.J.A.; Beerling, E.; Loomans, C.; Zomer, A.; Gerlach, C.; Vriekoop, N.; Seinstra, D.; Van Gurp, L.; Schafer, R.; et al. Intravital microscopy through an abdominal imaging window reveals a pre-micrometastasis stage during liver metastasis. Sci. Transl. Med. 2012, 4, 158ra145. [Google Scholar] [CrossRef] [PubMed]
- Ritsma, L.; Steller, E.J.A.; Ellenbroek, S.I.J.; Kranenburg, O.; Borel Rinkes, I.H.M.; Van Rheenen, J. Surgical implantation of an abdominal imaging window for intravital microscopy. Nat. Protoc. 2013, 8, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Bochner, F.; Fellus-Alyagor, L.; Kalchenko, V.; Shinar, S.; Neeman, M. A novel intravital imaging window for longitudinal microscopy of the mouse ovary. Sci. Rep. 2015, 5, 12446. [Google Scholar] [CrossRef] [PubMed]
- Yano, S.; Zhang, Y.; Miwa, S.; Tome, Y.; Hiroshima, Y.; Uehara, F.; Yamamoto, M.; Suetsugu, A.; Kishimoto, H.; Tazawa, H.; et al. Spatial-temporal FUCCI imaging of each cell in a tumor demonstrates locational dependence of cell cycle dynamics and chemoresponsiveness. Cell Cycle 2014, 13, 2110–2119. [Google Scholar] [CrossRef] [PubMed]
- Marusyk, A.; Almendro, V.; Polyak, K. Intra-tumour heterogeneity: A looking glass for cancer? Nat. Rev. Cancer 2012, 12, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Garcia, I.; Sole, R.V.; Costa, J. Metapopulation dynamics and spatial heterogeneity in cancer. Proc. Natl Acad. Sci. 2002, 99, 13085–13089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, R.K. Nomalization of Tumor Vasculature: An Emerging Concept in Antiangiogenic Therapy. Science 2005, 307, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Kienast, Y.; von Baumgarten, L.; Fuhrmann, M.; Klinkert, W.E.; Goldbrunner, R.; Herms, J.; Winkler, F. Real-time imaging reveals the single steps of brain metastasis formation. Nat. Med. 2010, 16, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Yano, S.; Takehara, K.; Tazawa, H.; Kishimoto, H.; Urata, Y.; Kagawa, S.; Fujiwara, T.; Hoffman, R.M. Cell-cycle-dependent drug-resistant quiescent cancer cells induce tumor angiogenesis after chemotherapy as visualized by real-time FUCCI imaging. Cell Cycle 2017, 16, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.D. Curing “incurable” cancer. Cancer Discov. 2011, 1, 477–480. [Google Scholar] [CrossRef] [PubMed]
- Gottesman, M.M. Mechanisms of cancer drug resistance. Annu. Rev. Med. 2002, 53, 615–627. [Google Scholar] [CrossRef] [PubMed]
- Aguirre-Ghiso, J.A. Models, mechanisms and clinical evidence for cancer dormancy. Nat. Rev. Cancer 2007, 7, 834–846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goss, P.E.; Chambers, A.F. Does tumour dormancy offer a therapeutic target? Nat. Rev. Cancer 2010, 10, 871–877. [Google Scholar] [CrossRef] [PubMed]
- Aguirre-Ghiso, J.A.; Bragado, P.; Sosa, M.S. Metastasis Awakening: Targeting dormant cancer. Nat. Med. 2013, 19, 276–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polzer, B.; Klein, C.A. The challenges of targeting minimal residual cancer. Nat. Med. 2013, 19, 274–275. [Google Scholar] [CrossRef] [PubMed]
- Kreso, A.; O’Brien, C.A.; van Galen, P.; Gan, O.I.; Notta, F.; Brown, A.M.; Ng, K.; Ma, J.; Wienholds, E.; Dunant, C.; et al. Viable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science 2013, 339, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, F.; Yoshida, S.; Saito, Y.; Hijikata, A.; Kitamura, H.; Tanaka, S.; Nakamura, R.; Tanaka, T.; Tomiyama, H.; Saito, N.; et al. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat. Biotechnol. 2007, 25, 1315–1321. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Kitamura, H.; Hijikata, A.; Tomizawa-Murasawa, M.; Tanaka, S.; Takagi, S.; Uchida, N.; Suzuki, N.; Sone, A.; Najima, Y.; et al. Identification of therapeutic targets for quiescent, chemotherapy-resistant human leukemia stem cells. Sci. Translational Med. 2010, 2, 17ra9. [Google Scholar] [CrossRef] [PubMed]
- Nakasone, E.S.; Askautrud, H.A.; Kees, T.; Park, J.H.; Plaks, V.; Ewald, A.J.; Fein, M.; Rasch, M.G.; Tan, Y.X.; Qiu, J.; et al. Imaging tumor-stroma interaction during chemotherapy reveals contributions of the microenvironment to resistance. Cancer Cell 2012, 12, 488–503. [Google Scholar] [CrossRef] [PubMed]
- Giedt, R. J.; Koch, P.D.; Weissleder, R. Single cell analysis of drug distribution by intravital imaging. PLoS ONE 2013, 8, e60988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conway, J.R.W.; Carragher, D.O.; Timpson, P. Developments in preclinical cancer imaging: Innovating the discovery of therapeutics. Nat. Rev. Cancer 2014, 14, 314–328. [Google Scholar] [CrossRef] [PubMed]
- Dan, S.; Okamura, M.; Mukai, Y.; Yoshimi, H.; Inoue, Y.; Hanyu, A.; Sakaue-Sawano, A.; Imamura, T.; Miyawaki, A.; Yamori, T. ZSTK474, a specific phosphatidylinositol 3-kinase inhibitor, induces G1 arrest of the cell cycle in vivo. Eur. J. Cancer. 2012, 48, 936–943. [Google Scholar] [CrossRef] [PubMed]
- Thurber, G.M.; Yang, K.S.; Reiner, T.; Kohler, R.H.; Sorger, P.; Mitchison, T.; Weissleder, R. Single-cell and subcellular pharmacokinetic imaging allows insight into drug action in vivo. Nat. Commun. 2013, 4, 1504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, Y.; Mukai, K.; Hishiki, T.; Kuo, A.; Ohmura, M.; Sugiura, Y.; Matsuura, T.; Nagahata, Y.; Hayakawa, N.; Yamamoto, T.; et al. Energy management by enhanced glycolysis in G1-phase in human colon cancer cells in vitro and in vivo. Mol. Cancer Res. 2013, 11, 973–985. [Google Scholar] [CrossRef] [PubMed]
- Haass, N.K.; Beaumont, K.A.; Hill, D.S.; Anfosso, A.; Mrass, P.; Munoz, M.A.; Kinjo, I.; Weninger, W. Real-time cell cycle imaging during melanoma growth, invasion, and drug response. Pigm. Cell Melanoma Res. 2014, 5, 764–776. [Google Scholar] [CrossRef] [PubMed]
- Orth, J.D.; Kohler, R.H.; Foijer, F.; Sorger, P.K.; Weissleder, R.; Mitchison, T.J. Analysis of Mitosis and Antimitotic Drug Responses in Tumors by In Vivo Microscopy and Single-Cell Pharmacodynamics. Cancer Res. 2011, 71, 4608–4616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janssen, A.; Beerling, E.; Medema, R.; van Rheenen, J. Intravital FRET imaging of tumor cell viability and mitosis during chemotherapy. PLoS ONE 2013, 8, e64029. [Google Scholar] [CrossRef] [PubMed]
- Zasadil, L.M.; Andersen, K.A.; Yeum, D.; Rocque, G.B.; Wilke, L.G.; Tevaarwerk, A.J.; Burkard, M.E.; Weaver, B.A. Cytotoxicity of paclitaxel in breast cancer is due to chromosome missegregation on multipolar spindles. Sci. Translational Med. 2014, 6, 229ra43. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, G.; Bouvette, G.; Therriault, H.; Bujold, R.; Saucier, C.; Paquette, B. Pre-irradiation of mouse mammary gland stimulates cancer cell migration and development of lung metastases. Br. J. Cancer. 2013, 109, 1829–1838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onozato, Y.; Kaida, A.; Harada, H.; Miura, M. Radiosensitivity of quiescent and proliferating cells grown as multicellular tumor spheroids. Cancer Sci. 2017, 108, 704–712. [Google Scholar] [CrossRef] [PubMed]
- Kagawa, Y.; Matsumoto, S.; Kamioka, Y.; Mimori, K.; Naito, Y.; Ishii, T.; Okuzaki, D.; Nishida, N.; Maeda, S.; Naito, A.; et al. Cell cycle-dependent Rho GTPase activity dynamically regulates cancer cell motility and invasion in vivo. PLoS ONE 2013, 8, e83629. [Google Scholar] [CrossRef] [PubMed]
- Yano, S.; Tazawa, H.; Hashimoto, Y.; Shirakawa, Y.; Kuroda, S.; Nishizaki, M.; Kishimoto, H.; Uno, F.; Nagasaka, T.; Urata, Y.; et al. A genetically engineered oncolytic adenovirus decoys and lethally traps quiescent cancer stem-like cells in S/G2/M phases. Clin. Cancer Res. 2013, 19, 6495–6505. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, T.; Kagawa, S.; Kobayashi, N.; Shirakiya, Y.; Umeoka, T.; Teraishi, F.; Taki, M.; Kyo, S.; Tanaka, N.; Fujiwara, T. Telomerase-specific replication-selective virotherapy for human cancer. Clin. Cancer Res. 2004, 10, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Nemunaitis, J.; Tong, A.W.; Nemunaitis, M.; Senzer, N.; Phadke, A.P.; Bedell, C.; Adams, N.; Zhang, Y.A.; Maples, P.B.; Chen, S.; et al. A phase I study of telomerase-specific replication competent oncolytic adenovirus (telomelysin) for various solid tumors. Mol. Ther. 2010, 18, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Yano, S.; Li, S.; Han, Q.; Tan, Y.; Bouvet, M.; Fujiwara, T.; Hoffman, R.M. Selective methioninase-induced trap of cancer cells in S/G2 phase visualized by FUCCI imaging confers chemosensitivity. Oncotarget 2014, 5, 8729–8736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yano, S.; Zhang, Y.; Zhao, M.; Hiroshima, Y.; Miwa, S.; Uehara, F.; Kishimoto, H.; Tazawa, H.; Bouvet, M.; Fujiwara, T.; et al. Tumor-targeting Salmonella typhimurium A1-R decoys quiescent cancer cells to cycle as visualized by FUCCI imaging and become sensitive to chemotherapy. Cell Cycle 2014, 13, 3958–3963. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Yang, M.; Li, X.M.; Jiang, P.; Baranov, E.; Li, S.; Xu, M.; Penman, S.; Hoffman, R.M. Tumor-targeting bacterial therapy with amino acid auxotrophs of GFP-expressing Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 2005, 102, 755–760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yano, S.; Takehara, K.; Zhao, M.; Tan, Y.; Han, Q.; Li, S.; Bouvet, M.; Fujiwara, T.; Hoffman, R.M. Tumor-specific cell-cycle decoy by Salmonella typhimurium A1-R combined with tumor-selective cell-cycle trap by methioninase overcome tumor intrinsic chemoresistance as visualized by FUCCI imaging. Cell Cycle 2016, 15, 1715–1723. [Google Scholar] [CrossRef] [PubMed] [Green Version]







© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yano, S.; Hoffman, R.M. Real-Time Determination of the Cell-Cycle Position of Individual Cells within Live Tumors Using FUCCI Cell-Cycle Imaging. Cells 2018, 7, 168. https://doi.org/10.3390/cells7100168
Yano S, Hoffman RM. Real-Time Determination of the Cell-Cycle Position of Individual Cells within Live Tumors Using FUCCI Cell-Cycle Imaging. Cells. 2018; 7(10):168. https://doi.org/10.3390/cells7100168
Chicago/Turabian StyleYano, Shuya, and Robert M. Hoffman. 2018. "Real-Time Determination of the Cell-Cycle Position of Individual Cells within Live Tumors Using FUCCI Cell-Cycle Imaging" Cells 7, no. 10: 168. https://doi.org/10.3390/cells7100168
APA StyleYano, S., & Hoffman, R. M. (2018). Real-Time Determination of the Cell-Cycle Position of Individual Cells within Live Tumors Using FUCCI Cell-Cycle Imaging. Cells, 7(10), 168. https://doi.org/10.3390/cells7100168