Nanopipettes as Monitoring Probes for the Single Living Cell: State of the Art and Future Directions in Molecular Biology
Abstract
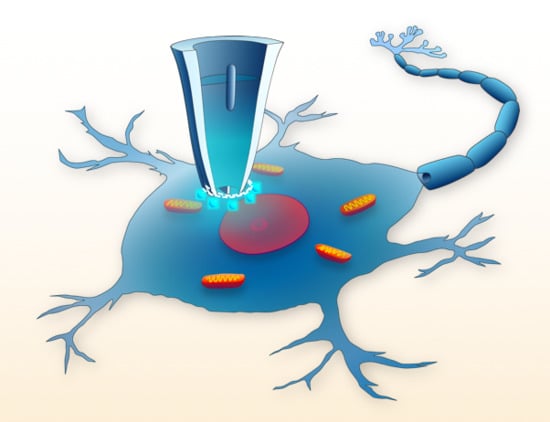
:1. Introduction
2. Use of Nanopipettes as Surgical Tools
2.1. Nanoinjections by Single-Cell Surgery
Intracellular Tracking of Injected Components
2.2. Single-Cell Nanobiopsy Platform
2.2.1. Single Cell Immunoassay
2.2.2. Genomics
2.2.3. Single Cell Aspiration
2.2.4. Nanogenomics
3. Monitoring Intracellular Components by Using Nanopipettes: Sensing
3.1. Layer-by-Layer (LbL) Immobilization of Recognition Elements
3.2. Electrochemical Techniques Used for Analysis
3.3. Recognition Element Selection for Immobilization on Nanopipettes
3.4. Specific Examples from the Literature
3.4.1. Glucose
3.4.2. pH and Reactive Oxygen Species
3.4.3. DNA
3.4.4. Metal Ions
4. Conclusions and Future Perspectives
Acknowledgments
Conflicts of Interest
References
- Sutter, P.W.; Sutter, E.A. Dispensing and surface-induced crystallization of zeptolitre liquid metal-alloy drops. Nat. Mater. 2007, 6, 363–366. [Google Scholar] [CrossRef] [PubMed]
- Laforge, F.O.; Carpino, J.; Rotenberg, S.A.; Mirkin, M.V. Electrochemical attosyringe. Proc. Natl. Acad. Sci. USA 2007, 104, 11895–11900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuoka, H.; Komazaki, T.; Mukai, Y.; Shibusawa, M.; Akane, H.; Chaki, A.; Uetake, N.; Saito, M. High throughput easy microinjection with a single-cell manipulation supporting robot. J. Biotechnol. 2005, 116, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Treutlein, B.; Brownfield, D.G.; Wu, A.R.; Neff, N.F.; Mantalas, G.L.; Espinoza, F.H.; Desai, T.J.; Krasnow, M.A.; Quake, S.R. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 2014, 509, 371–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Streets, A.M.; Zhang, X.; Cao, C.; Pang, Y.; Wu, X.; Xiong, L.; Yang, L.; Fu, Y.; Zhao, L.; Tang, F.; Huang, Y. Microfluidic single-cell whole-transcriptome sequencing. Proc. Natl. Acad. Sci. USA 2014, 111, 7048–7053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uehara, H.; Osada, T.; Ikai, A. Quantitative measurement of mRNA at different loci within an individual living cell. Ultramicroscopy 2004, 100, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-C.W.; Lopez-Diaz, F.J.; Khan, S.Y.; Tariq, M.A.; Dayn, Y.; Vaske, C.J.; Radenbaugh, A.J.; Kim, H.J.; Emerson, B.M.; Pourmand, N. Single-cell analyses of transcriptional heterogeneity during drug tolerance transition in cancer cells by RNA sequencing. Proc. Natl. Acad. Sci. USA 2014, 111, E4726–E4735. [Google Scholar] [CrossRef] [PubMed]
- Chaves, G.; Özel, R.E.; Rao, N.V.; Hadiprodjo, H.; Da Costa, Y.; Tokuno, Z.; Pourmand, N. Metabolic and transcriptomic analysis of Huntington’s disease model reveal changes in intracellular glucose levels and related genes. Heliyon 2017, 3, e00381. [Google Scholar] [CrossRef] [PubMed]
- Actis, P.; Mak, A.C.; Pourmand, N. Functionalized nanopipettes: Toward label-free, single cell biosensors. Bioanal. Rev. 2010, 1, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Karhanek, M.; Kemp, J.T.; Pourmand, N.; Davis, R.W.; Webb, C.D. Single DNA molecule detection using nanopipettes and nanoparticles. Single DNA molecule detection using nanopipettes and nanoparticles. Nano Lett. 2005, 5, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Actis, P.; Maalouf, M.M.; Kim, H.J.; Lohith, A.; Vilozny, B.; Seger, R.A.; Pourmand, N. Compartmental genomics in living cells revealed by single-cell nanobiopsy. ACS Nano 2013, 8, 546–553. [Google Scholar] [CrossRef] [PubMed]
- Vilozny, B.; Actis, P.; Seger, R.A.; Pourmand, N. Dynamic control of nanoprecipitation in a nanopipette. ACS Nano 2011, 5, 3191–3197. [Google Scholar] [CrossRef] [PubMed]
- Ying, L. Applications of Nanopipettes in Bionanotechnology; Portland Press Limited: London, UK, 2009. [Google Scholar]
- Morris, C.A.; Friedman, A.K.; Baker, L.A. Applications of nanopipettes in the analytical sciences. Analyst 2010, 135, 2190–2202. [Google Scholar] [CrossRef] [PubMed]
- Takami, T.; Park, B.H.; Kawai, T. Nanopipette exploring nanoworld. Nano Converg. 2014, 1, 17. [Google Scholar] [CrossRef] [PubMed]
- Neher, E.; Sakmann, B. Single-channel currents recorded from membrane of denervated frog muscle fibres. Nature 1976, 260, 799–802. [Google Scholar] [CrossRef] [PubMed]
- Mirkin, M.; Amemiya, S.; Mirkin, M. Nanoelectrodes and liquid/liquid nanointerfaces. In Nanoelectrochemistry; Mirkin, M.V., Amemiya, S., Eds.; CRC Press: Boca Raton, FL, USA, 2015; pp. 539–572. [Google Scholar]
- Trapnell, C.; Cacchiarelli, D.; Grimsby, J.; Pokharel, P.; Li, S.; Morse, M.; Lennon, N.J.; Livak, K.J.; Mikkelsen, T.S.; Rinn, J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014, 32, 381–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spitzer, M.H.; Nolan, G. Mass Cytometry: Single Cells, Many Features. Cell 2016, 165, 780–791. [Google Scholar] [CrossRef] [PubMed]
- Stephens, D.J.; Pepperkok, R. The many ways to cross the plasma membrane. Proc. Natl. Acad. Sci. USA 2001, 98, 4295–4298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meister, A.; Gabi, M.; Behr, P.; Studer, P.; Vörös, J.; Niedermann, P.; Bitterli, J.; Polesel-Maris, J.; Liley, M.; Heinzelmann, H.; et al. FluidFM: Combining Atomic Force Microscopy and Nanofluidics in a Universal Liquid Delivery System for Single Cell Applications and Beyond. Nano Lett. 2009, 9, 2501–2507. [Google Scholar] [CrossRef] [PubMed]
- Rodolfa, K.T.; Bruckbauer, A.; Zhou, D.; Korchev, Y.E.; Klenerman, D. Two-Component Graded Deposition of Biomolecules with a Double-Barreled Nanopipette. Angew. Chem. Int. Ed. 2005, 44, 6854–6859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.-C.; Wu, T.-H.; Clemens, D.L.; Lee, B.-Y.; Wen, X.; Horwitz, M.A.; Teitell, M.A.; Chiou, P.-Y. Massively parallel delivery of large cargo into mammalian cells with light pulses. Nat. Methods 2015, 12, 439–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, T.-H.; Sagullo, E.; Case, D.; Zheng, X.; Li, Y.; Hong, J.S.; TeSlaa, T.; Patananan, A.N.; McCaffery, J.M.; Niazi, K.; et al. Mitochondrial Transfer by Photothermal Nanoblade Restores Metabolite Profile in Mammalian Cells. Cell Metab. 2016, 23, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Tao, Y.; Lee, D.-H.; Wickramasinghe, H.K.; Lee, A.P. In situ mRNA isolation from a microfluidic single-cell array using an external AFM nanoprobe. Lab Chip 2017, 17, 1635–1644. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-C.; Baac, H.W.; Lee, K.-T.; Fouladdel, S.; Teichert, K.; Ok, J.G.; Cheng, Y.-H.; Ingram, P.N.; Hart, A.J.; Azizi, E. Selective Photomechanical Detachment and Retrieval of Divided Sister Cells from Enclosed Microfluidics for Downstream Analyses. ACS Nano 2017, 11, 4660–4668. [Google Scholar] [CrossRef] [PubMed]
- Seger, R.A.; Actis, P.; Penfold, C.; Maalouf, M.; Vilozny, B.; Pourmand, N. Voltage controlled nano-injection system for single-cell surgery. Nanoscale 2012, 4, 5843–5846. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Park, J.-H.; Choi, Y.; Heo, C.-J.; Yang, S.-M.; Lee, L.P.; Yang, P. Nanowire-based single-cell endoscopy. Nat. Nanotechnol. 2012, 7, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Singhal, R.; Orynbayeva, Z.; Sundaram, R.V.K.; Niu, J.J.; Bhattacharyya, S.; Vitol, E.A.; Schrlau, M.G.; Papazoglou, E.S.; Friedman, G.; Gogotsi, Y. Multifunctional carbon-nanotube cellular endoscopes. Nat. Nanotechnol. 2011, 6, 57–64. [Google Scholar] [CrossRef] [PubMed]
- König, J.; Zarnack, K.; Luscombe, N.M.; Ule, J. Protein–RNA interactions: New genomic technologies and perspectives. Nat. Rev. Genet. 2012, 13, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Chudakov, D.M.; Lukyanov, S.; Lukyanov, K.A. Fluorescent proteins as a toolkit for in vivo imaging. Trends Biotechnol. 2005, 23, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Voura, E.B.; Jaiswal, J.K.; Mattoussi, H.; Simon, S.M. Tracking metastatic tumor cell extravasation with quantum dot nanocrystals and fluorescence emission-scanning microscopy. Nat. Med. 2004, 10, 993–998. [Google Scholar] [CrossRef] [PubMed]
- Junker, J.P.; Spanjaard, B.; Peterson-Maduro, J.; Alemany, A.; Hu, B.; Florescu, M.; van Oudenaarden, A. Massively parallel clonal analysis using CRISPR/Cas9 induced genetic scars. bioRxiv 2017. [Google Scholar] [CrossRef]
- Kretzschmar, K.; Watt, F.M. Lineage tracing. Cell 2012, 148, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Kanisicak, O.; Khalil, H.; Ivey, M.J.; Karch, J.; Maliken, B.D.; Correll, R.N.; Brody, M.J.; Lin, S.-C.J.; Aronow, B.J.; Tallquist, M.D.; et al. Genetic lineage tracing defines myofibroblast origin and function in the injured heart. Nat. Commun. 2016, 7, 12260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beckman, R.A.; Schemmann, G.S.; Yeang, C.-H. Impact of genetic dynamics and single-cell heterogeneity on development of nonstandard personalized medicine strategies for cancer. Proc. Natl. Acad. Sci. USA 2012, 109, 14586–14591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S. Drug Resistance in Cancer: An Overview. Cancers 2014, 6, 1769–1792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alfarouk, K.O.; Stock, C.-M.; Taylor, S.; Walsh, M.; Muddathir, A.K.; Verduzco, D.; Bashir, A.H.; Mohammed, O.Y.; Elhassan, G.O.; Harguindey, S.; et al. Resistance to cancer chemotherapy: Failure in drug response from ADME to P-gp. Cancer Cell Int. 2015, 15, 71. [Google Scholar] [CrossRef] [PubMed]
- Komarova, N.L.; Wodarz, D. Drug resistance in cancer: Principles of emergence and prevention. Proc. Natl. Acad. Sci. USA 2005, 102, 9714–9719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sui, X.; Chen, R.; Wang, Z.; Huang, Z.; Kong, N.; Zhang, M.; Han, W.; Lou, F.; Yang, J.; Zhang, Q.; et al. Autophagy and chemotherapy resistance: A promising therapeutic target for cancer treatment. Cell Death Dis. 2013, 4, e838. [Google Scholar] [CrossRef] [PubMed]
- Borst, P. Cancer drug pan-resistance: Pumps, cancer stem cells, quiescence, epithelial to mesenchymal transition, blocked cell death pathways, persisters or what? Open Biol. 2012, 2, 120066. [Google Scholar] [CrossRef] [PubMed]
- Creighton, T.E. Protein Structure: A Practical Approach; Oxford University Press: Oxford, NY, USA, 1997. [Google Scholar]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Renart, J.; Reiser, J.; Stark, G.R. Transfer of proteins from gels to diazobenzyloxymethyl-paper and detection with antisera: A method for studying antibody specificity and antigen structure. Proc. Natl. Acad. Sci. USA 1979, 76, 3116–3120. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.J.; Spelke, D.P.; Xu, Z.; Kang, C.-C.; Schaffer, D.V.; Herr, A.E. Single-cell western blotting. Nat. Methods 2014, 11, 749–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, W.; Shin, Y.S.; Ma, C.; Wang, J.; Elitas, M.; Fan, R.; Heath, J.R. Microchip platforms for multiplex single-cell functional proteomics with applications to immunology and cancer research. Genome Med. 2013, 5, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Umehara, S.; Karhanek, M.; Davis, R.W.; Pourmand, N. Label-free biosensing with functionalized nanopipette probes. Proc. Natl. Acad. Sci. USA 2009, 106, 4611–4616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venter, J.C.; Adams, M.D.; Myers, E.W.; Li, P.W.; Mural, R.J.; Sutton, G.G.; Smith, H.O.; Yandell, M.; Evans, C.A.; Holt, R.A.; et al. The Sequence of the Human Genome. Science 2001, 291, 1304–1351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, J.W.; Locke, J.C.W.; Altinok, A.; Rosenfeld, N.; Bacarian, T.; Swain, P.S.; Mjolsness, E.; Elowitz, M.B. Measuring single-cell gene expression dynamics in bacteria using fluorescence time-lapse microscopy. Nat. Protoc. 2012, 7, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Glauche, I.; Herberg, M.; Roeder, I. Nanog variability and pluripotency regulation of embryonic stem cells-insights from a mathematical model analysis. PLoS ONE 2010, 5, e11238. [Google Scholar] [CrossRef] [PubMed]
- Dale, S.E.C.; Unwin, P.R. Polarised liquid/liquid micro-interfaces move during charge transfer. Electrochem. Commun. 2008, 10, 723–726. [Google Scholar] [CrossRef]
- Tóth, E.N.; Lohith, A.; Mondal, M.; Guo, J.; Fukamizu, A.; Pourmand, N. Single-cell nanobiopsy reveals compartmentalization of mRNA in neuronal cells. J. Biol. Chem. 2018, 293, 4940–4951. [Google Scholar] [CrossRef] [PubMed]
- Nashimoto, Y.; Takahashi, Y.; Zhou, Y.; Ito, H.; Ida, H.; Ino, K.; Matsue, T.; Shiku, H. Evaluation of mRNA Localization Using Double Barrel Scanning Ion Conductance Microscopy. ACS Nano 2016, 10, 6915–6922. [Google Scholar] [CrossRef] [PubMed]
- Guillaume-Gentil, O.; Grindberg, R.V.; Kooger, R.; Dorwling-Carter, L.; Martinez, V.; Ossola, D.; Pilhofer, M.; Zambelli, T.; Vorholt, J.A. Tunable Single-Cell Extraction for Molecular Analyses. Cell 2016, 166, 506–516. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Li, L. Metabolomics of Small Numbers of Cells: Metabolomic Profiling of 100, 1000, and 10000 Human Breast Cancer Cells. Anal. Chem. 2017, 89, 11664–11671. [Google Scholar] [CrossRef] [PubMed]
- Guillaume-Gentil, O.; Rey, T.; Kiefer, P.; Ibáñez, A.J.; Steinhoff, R.; Brönnimann, R.; Dorwling-Carter, L.; Zambelli, T.; Zenobi, R.; Vorholt, J.A. Single-Cell Mass Spectrometry of Metabolites Extracted from Live Cells by Fluidic Force Microscopy. Anal. Chem. 2017, 89, 5017–5023. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Hjort, M.; Chen, H.; Birey, F.; Leal-Ortiz, S.A.; Han, C.M.; Santiago, J.G.; Paşca, S.P.; Wu, J.C.; Melosh, N.A. Nondestructive nanostraw intracellular sampling for longitudinal cell monitoring. Proc. Natl. Acad. Sci. USA 2017, 114, E1866–E1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nawarathna, D.; Chang, R.; Nelson, E.; Wickramasinghe, H.K. Targeted mRNA Profiling of Transfected Breast Cancer Gene in a Living Cell. Anal. Biochem. 2011, 408, 342–344. [Google Scholar] [CrossRef] [PubMed]
- Uehara, H.; Kunitomi, Y.; Ikai, A.; Osada, T. mRNA detection of individual cells with the single cell nanoprobe method compared with in situ hybridization. J. Nanobiotechnol. 2007, 5, 7. [Google Scholar] [CrossRef] [PubMed]
- Clark, I.E.; Dodson, M.W.; Jiang, C.; Cao, J.H.; Huh, J.R.; Seol, J.H.; Yoo, S.J.; Hay, B.A.; Guo, M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 2006, 441, 1162–1166. [Google Scholar] [CrossRef] [PubMed]
- Ståhlberg, A.; Thomsen, C.; Ruff, D.; Åman, P. Quantitative PCR analysis of DNA, RNAs, and proteins in the same single cell. Clin. Chem. 2012, 58, 1682–1691. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Schlesinger, F.; Davis, C.A.; Zhang, Y.; Li, R.; Salit, M.; Gingeras, T.R.; Oliver, B. Synthetic spike-in standards for RNA-seq experiments. Genome Res. 2011, 21, 1543–1551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klijn, C.; Durinck, S.; Stawiski, E.W.; Haverty, P.M.; Jiang, Z.; Liu, H.; Degenhardt, J.; Mayba, O.; Gnad, F.; Liu, J.; et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat. Biotechnol. 2015, 33, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Ramdas, P.; Rajihuzzaman, M.; Veerasenan, S.D.; Selvaduray, K.R.; Nesaretnam, K.; Radhakrishnan, A.K. Tocotrienol-treated MCF-7 human breast cancer cells show down-regulation of API5 and up-regulation of MIG6 genes. Cancer Genom. Proteom. 2011, 8, 19–31. [Google Scholar]
- Lyng, M.B.; Lænkholm, A.-V.; Pallisgaard, N.; Ditzel, H.J. Identification of genes for normalization of real-time RT-PCR data in breast carcinomas. BMC Cancer 2008, 8, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shadeo, A.; Lam, W.L. Comprehensive copy number profiles of breast cancer cell model genomes. Breast Cancer Res. 2006, 8, R9. [Google Scholar] [CrossRef] [PubMed]
- Linger, R.M.; Keating, A.K.; Earp, H.S.; Graham, D.K. TAM receptor tyrosine kinases: Biologic functions, signaling, and potential therapeutic targeting in human cancer. Adv. Cancer Res. 2008, 100, 35–83. [Google Scholar] [PubMed]
- Zhang, X.; Schulz, R.; Edmunds, S.; Krüger, E.; Markert, E.; Gaedcke, J.; Cormet-Boyaka, E.; Ghadimi, M.; Beissbarth, T.; Levine, A.J.; et al. MicroRNA-101 suppresses tumor cell proliferation by acting as an endogenous proteasome inhibitor via targeting the proteasome assembly factor POMP. Mol. Cell 2015, 59, 243–257. [Google Scholar] [CrossRef] [PubMed]
- Gorelik, J.; Gu, Y.; Spohr, H.A.; Shevchuk, A.I.; Lab, M.J.; Harding, S.E.; Edwards, C.R.W.; Whitaker, M.; Moss, G.W.J.; Benton, D.C.H.; et al. Ion Channels in Small Cells and Subcellular Structures Can Be Studied with a Smart Patch-Clamp System. Biophys. J. 2002, 83, 3296–3303. [Google Scholar] [CrossRef] [Green Version]
- Svensson, K.; Olin, H.; Olsson, E. Nanopipettes for metal transport. Phys. Rev. Lett. 2004, 93, 145901. [Google Scholar] [CrossRef] [PubMed]
- Ying, L.; Bruckbauer, A.; Zhou, D.; Gorelik, J.; Shevchuk, A.; Korchev, Y.; Klenerman, D. The scanned nanopipette: A new tool for high resolution bioimaging and controlled deposition of biomolecules. Phys. Chem. Chem. Phys. 2005, 7, 2859–2866. [Google Scholar] [CrossRef] [PubMed]
- Umehara, S.; Pourmand, N.; Webb, C.D.; Davis, R.W.; Yasuda, K.; Karhanek, M. Current rectification with poly-l-lysine-coated quartz nanopipettes. Nano Lett. 2006, 6, 2486–2492. [Google Scholar] [CrossRef] [PubMed]
- Rodolfa, K.T.; Bruckbauer, A.; Zhou, D.; Schevchuk, A.I.; Korchev, Y.E.; Klenerman, D. Nanoscale Pipetting for Controlled Chemistry in Small Arrayed Water Droplets Using a Double-Barrel Pipet. Nano Lett. 2006, 6, 252–257. [Google Scholar] [CrossRef] [PubMed]
- Iwata, F.; Nagami, S.; Sumiya, Y.; Sasaki, A. Nanometre-scale deposition of colloidal Au particles using electrophoresis in a nanopipette probe. Nanotechnology 2007, 18, 105301. [Google Scholar] [CrossRef]
- Bruckbauer, A.; James, P.; Zhou, D.; Yoon, J.W.; Excell, D.; Korchev, Y.; Jones, R.; Klenerman, D. Nanopipette Delivery of Individual Molecules to Cellular Compartments for Single-Molecule Fluorescence Tracking. Biophys. J. 2007, 93, 3120–3131. [Google Scholar] [CrossRef] [PubMed]
- Byun, C.K.; Wang, X.; Pu, Q.; Liu, S. Electroosmosis-Based Nanopipettor. Anal. Chem. 2007, 79, 3862–3866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karhanek, M.; Webb, C.D.; Umehara, S.; Pourmand, N. Functionalized Nanopipette Biosensor. U.S. Patent Application No. 20100072080, 25 March 2010. [Google Scholar]
- Rothery, A.; Gorelik, J.; Bruckbauer, A.; Yu, W.; Korchev, Y.; Klenerman, D. A novel light source for SICM–SNOM of living cells. J. Microsc. 2003, 209, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Gorelik, J.; Zhang, Y.; Shevchuk, A.I.; Frolenkov, G.I.; Sánchez, D.; Vodyanoy, I.; Edwards, C.R.; Klenerman, D.; Korchev, Y.E. The use of scanning ion conductance microscopy to image A6 cells. Mol. Cell. Endocrinol. 2004, 217, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Windhorst, U.; Johansson, H. Modern Techniques in Neuroscience Research; Springer Science & Business Media: Berlin, Germany, 2012. [Google Scholar]
- Ying, L.; White, S.S.; Bruckbauer, A.; Meadows, L.; Korchev, Y.E.; Klenerman, D. Frequency and Voltage Dependence of the Dielectrophoretic Trapping of Short Lengths of DNA and dCTP in a Nanopipette. Biophys. J. 2004, 86, 1018–1027. [Google Scholar] [CrossRef] [Green Version]
- Ying, L.; Bruckbauer, A.; Rothery, A.M.; Korchev, Y.E.; Klenerman, D. Programmable Delivery of DNA through a Nanopipet. Anal. Chem. 2002, 74, 1380–1385. [Google Scholar] [CrossRef] [PubMed]
- Clarke, R.W.; White, S.S.; Zhou, D.; Ying, L.; Klenerman, D. Trapping of proteins under physiological conditions in a nanopipette. Angew. Chem. Int. Ed. 2005, 44, 3747–3750. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.-J.; Ying, Y.-L.; Hu, Y.-X.; Gao, R.; Long, Y.-T. Label-free monitoring of single molecule immunoreaction with a nanopipette. Anal. Chem. 2017, 89, 8203–8206. [Google Scholar] [CrossRef] [PubMed]
- Chambers, J.P.; Arulanandam, B.P.; Matta, L.L.; Weis, A.; Valdes, J.J. Biosensor Recognition Elements; Department of Biology, University of Texas at San Antonio: San Antonio, TX, USA, 2008. [Google Scholar]
- Thiviyanathan, V.; Gorenstein, D.G. Aptamers and the next generation of diagnostic reagents. Proteom. Clin. Appl. 2012, 6, 563–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, A.Z.; Farokhzad, O.C. Current progress of aptamer-based molecular imaging. J. Nucl. Med. 2014, 55, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Actis, P.; Rogers, A.; Nivala, J.; Vilozny, B.; Seger, R.A.; Jejelowo, O.; Pourmand, N. Reversible thrombin detection by aptamer functionalized STING sensors. Biosens. Bioelectron. 2011, 26, 4503–4507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nascimento, R.A.; Özel, R.E.; Mak, W.H.; Mulato, M.; Singaram, B.; Pourmand, N. Single cell ‘glucose nanosensor’ verifies elevated glucose levels in individual cancer cells. Nano Lett. 2016, 16, 1194–1200. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Annibaldi, A.; Widmann, C. Glucose metabolism in cancer cells. Curr. Opin. Clin. Nutr. Metab. Care 2010, 13, 466–470. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Sempos, C.T.; Freudenheim, J.L.; Muti, P.; Smit, E. Serum iron, copper and zinc concentrations and risk of cancer mortality in US adults. Ann. Epidemiol. 2004, 14, 195–201. [Google Scholar] [CrossRef]
- Wiseman, H.; Halliwell, B. Damage to DNA by reactive oxygen and nitrogen species: Role in inflammatory disease and progression to cancer. Biochem. J. 1996, 313 Pt 1, 17. [Google Scholar] [CrossRef] [PubMed]
- Gatenby, R.A.; Gillies, R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 2004, 4, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Özel, R.E.; Lohith, A.; Mak, W.H.; Pourmand, N. Single-cell intracellular nano-pH probes. RSC Adv. 2015, 5, 52436–52443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azad, N.; Rojanasakul, Y.; Vallyathan, V. Inflammation and lung cancer: Roles of reactive oxygen/nitrogen species. J. Toxicol. Environ. Health Part B 2008, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Winterbourn, C.C. The challenges of using fluorescent probes to detect and quantify specific reactive oxygen species in living cells. Biochim. Biophys. Acta (BBA) Gen. Subj. 2014, 1840, 730–738. [Google Scholar] [CrossRef] [PubMed]
- Steinbock, L.J.; Otto, O.; Chimerel, C.; Gornall, J.; Keyser, U.F. Detecting DNA Folding with Nanocapillaries. Nano Lett. 2010, 10, 2493–2497. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Patil, A.V.; Ivanov, A.P.; Kong, Q.; Gibb, T.; Dogan, F.; deMello, A.J.; Edel, J.B. Label-Free In-Flow Detection of Single DNA Molecules using Glass Nanopipettes. Anal. Chem. 2014, 86, 835–841. [Google Scholar] [CrossRef] [PubMed]
- Actis, P.; Vilozny, B.; Seger, R.A.; Li, X.; Jejelowo, O.; Rinaudo, M.; Pourmand, N. Voltage-controlled metal binding on polyelectrolyte-functionalized nanopores. Langmuir 2011, 27, 6528–6533. [Google Scholar] [CrossRef] [PubMed]
- Vilozny, B.; Actis, P.; Seger, R.A.; Vallmajo-Martin, Q.; Pourmand, N. Reversible Cation Response with a Protein-Modified Nanopipette. Anal. Chem. 2011, 83, 6121–6126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sa, N.; Fu, Y.; Baker, L.A. Reversible Cobalt Ion Binding to Imidazole-Modified Nanopipettes. Anal. Chem. 2010, 82, 9963–9966. [Google Scholar] [CrossRef] [PubMed] [Green Version]









| RefSeq mRNA Accession | Gene Symbol | Gene Name |
|---|---|---|
| NM_001001521 | UGP2 | UDP-glucose pyrophosphorylase |
| NM_001002 | RPLP0 | ribosomal protein lateral stalk subunit P0 |
| NM_001017963 | HSP90AA1 | heat shock protein 90 α family class A member 1 |
| NM_001201483 | ENO1 | enolase 1 |
| NM_001402 | EEF1A1 | eukaryotic translation elongation factor 1 α 1 |
| NM_001699 | AXL | AXL receptor tyrosine kinase |
| NM_002520 | NPM1 | nucleophosmin 1 |
| NM_005324_mRNA | H3F3B | H3 histone, family 3B |
| NM_015932_mRNA | POMP | proteasome maturation protein |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bulbul, G.; Chaves, G.; Olivier, J.; Ozel, R.E.; Pourmand, N. Nanopipettes as Monitoring Probes for the Single Living Cell: State of the Art and Future Directions in Molecular Biology. Cells 2018, 7, 55. https://doi.org/10.3390/cells7060055
Bulbul G, Chaves G, Olivier J, Ozel RE, Pourmand N. Nanopipettes as Monitoring Probes for the Single Living Cell: State of the Art and Future Directions in Molecular Biology. Cells. 2018; 7(6):55. https://doi.org/10.3390/cells7060055
Chicago/Turabian StyleBulbul, Gonca, Gepoliano Chaves, Joseph Olivier, Rifat Emrah Ozel, and Nader Pourmand. 2018. "Nanopipettes as Monitoring Probes for the Single Living Cell: State of the Art and Future Directions in Molecular Biology" Cells 7, no. 6: 55. https://doi.org/10.3390/cells7060055
APA StyleBulbul, G., Chaves, G., Olivier, J., Ozel, R. E., & Pourmand, N. (2018). Nanopipettes as Monitoring Probes for the Single Living Cell: State of the Art and Future Directions in Molecular Biology. Cells, 7(6), 55. https://doi.org/10.3390/cells7060055