The Unfolded Protein Response Pathway in the Yeast Kluyveromyces lactis. A Comparative View among Yeast Species
Abstract
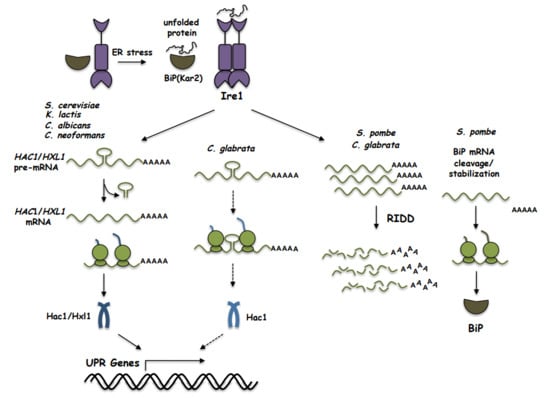
:1. Introduction
2. The UPR in Saccharomyces cerevisiae
3. The UPR in Kluyveromyces lactis
4. The UPR in Schizosaccharomyces pombe
5. The UPR in Candida glabrata
6. The UPR in Cryptococcus neoformans
7. The UPR in Candida albicans
8. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Domínguez-Martín, E.; Ongay-Larios, L.; Kawasaki, L.; Vincent, O.; Coello, G.; Coria, R.; Escalante, R. IreA controls endoplasmic reticulum stress-induced autophagy and survival through homeostasis recovery. Mol. Cell. Biol. 2018, 38, e00054-18. [Google Scholar] [CrossRef]
- Domínguez-Martín, E.; Hernández-Elvira, M.; Vincent, O.; Coria, R.; Escalante, R. Unfolding the endoplasmic reticulum of a social amoeba: Dictyostelium discoideum as a new model for the study of endoplasmic reticulum stress. Cells 2018, 7, 56. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Ma, W.; Gething, M.J.; Sambrook, J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell 1993, 74, 743–756. [Google Scholar] [CrossRef] [PubMed]
- Sidrauski, C.; Walter, P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 1997, 90, 1031–1039. [Google Scholar] [CrossRef]
- Bertolotti, A.; Zhang, Y.; Hendershot, L.M.; Harding, H.P.; Ron, D. Dynamic interaction of BiP and ER stress transducers in the unfolded- protein response. Nat. Cell Biol. 2000, 2, 326–332. [Google Scholar] [CrossRef] [PubMed]
- Okamura, K.; Kimata, Y.; Higashio, H.; Tsuru, A.; Kohno, K. Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem. Biophys. Res. Commun. 2000, 279, 445–450. [Google Scholar] [CrossRef] [PubMed]
- Credle, J.J.; Finer-Moore, J.S.; Papa, F.R.; Stroud, R.M.; Walter, P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2005, 102, 18773–18784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oikawa, D.; Kimata, Y.; Kohno, K. Self-association and BiP dissociation are not sufficient for activation of the ER stress sensor Ire1. J. Cell Sci. 2007, 120, 1681–1688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimata, Y.; Ishiwata-Kimata, Y.; Ito, T.; Hirata, A.; Suzuki, T.; Oikawa, D.; Takeuchi, M.; Kohno, K. Two regulatory steps of ER-stress sensor Ire1 involving its cluster formation and interaction with unfolded proteins. J. Cell Biol. 2007, 179, 75–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gardner, B.M.; Walter, P. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science 2011, 333, 1891–1894. [Google Scholar] [CrossRef] [PubMed]
- Nikawa, J.; Yamashita, S. IRE1 encodes a putative protein kinase containing a membrane-spanning domain and is required for inositol phototrophy in Saccharomyces cerevisiae. Mol. Microbiol. 1992, 6, 1441–1446. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.S.; Shamu, C.E.; Walter, P. Transcriptional induction of genes encoding endoplasmic-reticulum resident proteins requires a transmembrane protein-kinase. Cell 1993, 73, 1197–1206. [Google Scholar] [CrossRef]
- Sidrauski, C.; Cox, J.S.; Walter, P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell 1996, 87, 405–413. [Google Scholar] [CrossRef]
- Cox, J.S.; Chapman, R.E.; Walter, P. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol. Biol. Cell 1997, 8, 1805–1814. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y. Identification of mitogen-activated protein kinase signaling pathways that confer resistance to endoplasmic reticulum stress in Saccharomyces cerevisiae. Mol. Cancer Res. 2005, 3, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Shamu, C.E.; Walter, P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996, 15, 3028–3039. [Google Scholar] [PubMed]
- Van Anken, E.; Pincus, D.; Coyle, S.; Aragón, T.; Osman, C.; Lari, F.; Gómez Puerta, S.; Korennykh, A.V.; Walter, P. Specificity in endoplasmic reticulum-stress signaling in yeast entails a step-wise engagement of HAC1 mRNA to clusters of the stress sensor Ire1. eLife 2014, 3, e05031. [Google Scholar] [CrossRef] [PubMed]
- Goffin, L.; Vodala, S.; Fraser, C.; Ryan, J.; Timms, M.; Meusburger, S.; Catimel, B.; Nice, E.C.; Silver, P.A.; Xiao, C.-Y.; et al. The Unfolded Protein Response transducer Ire1p contains a nuclear localization sequence recognized by multiple β importins. Mol. Biol. Cell 2006, 17, 5309–5323. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Kawahara, T.; Yoshida, H.; Yanagi, H.; Yura, T. Signalling from endoplasmic reticulum to nucleus: Transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells 1996, 1, 803–817. [Google Scholar] [CrossRef] [PubMed]
- Fordyce, P.M.; Pincus, D.; Kimmig, P.; Nelson, C.S.; El-Samad, H.; Walter, P.; DeRisi, J.L. Basic leucine zipper transcription factor Hac1 binds DNA in two distinct modes as revealed by microfluidic analyses. Proc. Natl. Acad. Sci. USA 2012, 109, E3084–E3093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujii, Y.; Shimizu, T.; Toda, T.; Yanagida, M.; Hakoshima, T. Structural basis for the diversity of DNA recognition by bZIP transcription factors. Nat. Struct. Biol. 2000, 7, 889–893. [Google Scholar] [CrossRef] [PubMed]
- Miller, M. The importance of being flexible: The case of basic region leucine zipper transcriptional regulators. Curr. Protein Pept. Sci. 2009, 10, 244–269. [Google Scholar] [CrossRef] [PubMed]
- Nojima, H.; Leem, S.H.; Araki, H.; Sakai, A.; Nakashima, N.; Kanaoka, Y.; Ono, Y. Hac1: A novel yeast bZIP protein binding to the CRE motif is a multicopy suppressor for cdc10 mutant of Schizosaccharomyces pombe. Nucleic Acids Res. 1994, 22, 5279–5288. [Google Scholar] [CrossRef] [PubMed]
- Nikawa, J.I.; Akiyoshi, M.; Hirata, S.; Fukuda, T. Saccharomyces cerevisiae IRE2/HAC1 is involved in IRE1-mediated KAR2 expression. Nucleic Acids Res. 1996, 24, 4222–4226. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.S.; Walter, P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 1996, 87, 391–404. [Google Scholar] [CrossRef]
- Chapman, R.E.; Walter, P. Translational attenuation mediated by an mRNA intron. Curr. Biol. 1997, 7, 850–859. [Google Scholar] [CrossRef]
- Kawahara, T.; Yanagi, H.; Yura, T.; Mori, K. Endoplasmic reticulum stress-induced mRNA splicing permits synthesis of transcription factor Hac1p/Ern4p that activates the unfolded protein response. Mol. Biol. Cell 1997, 8, 1845–1862. [Google Scholar] [CrossRef] [PubMed]
- Rüegsegger, U.; Leber, J.H.; Walter, P. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell 2001, 107, 103–114. [Google Scholar] [CrossRef]
- Aragón, T.; Van Anken, E.; Pincus, D.; Serafimova, I.M.; Korennykh, A.V.; Rubio, C.A.; Walter, P. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature 2009, 457, 736–740. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Ogawa, N.; Kawahara, T.; Yanagi, H.; Yura, T. mRNA splicing-mediated C-terminal replacement of transcription factor Hac1p is required for efficient activation of the unfolded protein response. Proc. Natl. Acad. Sci. USA 2000, 97, 4660–4665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, K.; Sant, A.; Kohno, K.; Normington, K.; Gething, M.J.; Sambrook, J.F. A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2 (BiP) gene by unfolded proteins. EMBO J. 1992, 11, 2583–2593. [Google Scholar] [PubMed]
- Kohno, K.; Normington, K.; Sambrook, J.; Gething, M.J.; Mori, K. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol. Cell. Biol. 1993, 13, 877–890. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Ogawa, N.; Kawahara, T.; Yanagi, H.; Yura, T. Palindrome with spacer of one nucleotide is characteristic of the cis-acting unfolded protein response element in Saccharomyces cerevisiae. J. Biol. Chem. 1998, 273, 9912–9920. [Google Scholar] [CrossRef] [PubMed]
- Partaledis, J.A.; Berlin, V. The FKB2 gene of Saccharomyces cerevisiae, encoding the immunosuppressant-binding protein FKBP-13, is regulated in response to accumulation of unfolded proteins in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 1993, 90, 5450–5454. [Google Scholar] [CrossRef] [PubMed]
- Patil, C.K.; Li, H.; Walter, P. Gcn4p and novel upstream activating sequences regulate targets of the unfolded protein response. PLoS Biol. 2004, 2, e246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bukau, B.; Horwich, A.L. The Hsp70 and Hsp60 Chaperone Machines. Cell 1998, 92, 351–366. [Google Scholar] [CrossRef]
- Vogel, J.P.; Misra, L.M.; Rose, M.D. Loss of BiP/GRP78 function blocks translocation of secretory proteins in yeast. J. Cell Biol. 1990, 110, 1885–1895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishikawa, S.I.; Fewell, S.W.; Kato, Y.; Brodsky, J.L.; Endo, T. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J. Cell Biol. 2001, 153, 1061–1070. [Google Scholar] [CrossRef] [PubMed]
- Kimata, Y.; Kimata, Y.I.; Shimizu, Y.; Abe, H.; Farcasanu, I.C.; Takeuchi, M.; Rose, M.D.; Kohno, K. Genetic evidence for a role of BiP/Kar2 that regulates Ire1 in response to accumulation of unfolded proteins. Mol. Biol. Cell 2003, 14, 2559–2569. [Google Scholar] [CrossRef] [PubMed]
- Normington, K.; Kohno, K.; Kozutsumi, Y.; Gething, M.J.; Sambrook, J. S. cerevisiae encodes an essential protein homologous in sequence and function to mammalian BiP. Cell 1989, 57, 1223–1236. [Google Scholar] [CrossRef]
- Wolfe, K.H.; Shields, D.C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 1997, 387, 708–713. [Google Scholar] [CrossRef] [PubMed]
- Hooks, K.B.; Griffiths-Jones, S. Conserved RNA structures in the non-canonical Hac1/Xbp1 intron. RNA Biol. 2011, 8, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Mori, T.; Ogasawara, C.; Inada, T.; Englert, M.; Beier, H.; Takezawa, M.; Endo, T.; Yoshihisa, T. Dual functions of yeast tRNA ligase in the Unfolded Protein Response: Unconventional cytoplasmic splicing of HAC1 pre-mRNA is not sufficient to release translational attenuation. Mol. Biol. Cell 2010, 21, 3722–3734. [Google Scholar] [CrossRef] [PubMed]
- Lewis, M.J.; Pelham, H.R. The sequence of the Kluyveromyces lactis BiP gene. Nucleic Acids Res. 1990, 18, 6438. [Google Scholar] [CrossRef] [PubMed]
- Lewis, M.J.; Sweet, D.J.; Pelham, H.R. The ERD2 gene determines the specificity of the luminal ER protein retention system. Cell 1990, 61, 1359–1363. [Google Scholar] [CrossRef] [Green Version]
- Pidoux, A.L.; Armstrong, J. The BiP protein and the endoplasmic reticulum of Schizosaccharomyces pombe: Fate of the nuclear envelope during cell division. J. Cell. Sci. 1993, 105, 1115–1120. [Google Scholar] [PubMed]
- Frost, A.; Elgort, M.G.; Brandman, O.; Ives, C.; Collins, S.R.; Miller-Vedam, L.; Weibezahn, J.; Hein, M.Y.; Poser, I.; Mann, M.; et al. Functional repurposing revealed by comparing S. pombe and S. cerevisiae genetic interactions. Cell 2012, 149, 1339–1352. [Google Scholar] [CrossRef] [PubMed]
- Ridgway, G.J.; Douglas, H.C. Unbalanced growth of yeast due to inositol deficiency. J. Bacteriol. 1958, 76, 163–166. [Google Scholar] [PubMed]
- Ingavale, S.S.; Bachhawat, A.K. Restoration of inositol prototrophy in the fission yeast Schizosaccharomyces pombe. Microbiology 1999, 145, 1903–1910. [Google Scholar] [CrossRef] [PubMed]
- Voicu, P.-M.; Poitelea, M.; Schweingruber, E.; Rusu, M. Inositol is specifically involved in the sexual program of the fission yeast Schizosaccharomyces pombe. Arch. Microbiol. 2002, 177, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Niederberger, C.; Gräub, R.; Schweingruber, A.-M.; Fankhauser, H.; Rusu, M.; Poitelea, M.; Edenharter, L.; Schweingruber, M.E. Exogenous inositol and genes responsible for inositol transport are required for mating and sporulation in Schizosaccharomyces pombe. Curr. Genet. 1998, 33, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Kimmig, P.; Diaz, M.; Zheng, J.; Williams, C.C.; Lang, A.; Aragón, T.; Li, H.; Walter, P. The unfolded protein response in fission yeast modulates stability of select mRNAs to maintain protein homeostasis. eLife 2012, 2012, e00048. [Google Scholar] [CrossRef] [PubMed]
- Hollien, J.; Weissman, J.S. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 2006, 313, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Mishiba, K.; Nagashima, Y.; Suzuki, E.; Hayashi, N.; Ogata, Y.; Shimada, Y.; Koizumi, N. Defects in IRE1 enhance cell death and fail to degrade mRNAs encoding secretory pathway proteins in the Arabidopsis unfolded protein response. Proc. Natl. Acad. Sci. USA 2013, 110, 5713–5718. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Lerner, A.G.; Vande Walle, L.; Upton, J.-P.; Xu, W.; Hagen, A.; Backes, B.J.; Oakes, S.A.; Papa, F.R. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell 2009, 138, 562–575. [Google Scholar] [CrossRef] [PubMed]
- Hollien, J.; Lin, J.H.; Li, H.; Stevens, N.; Walter, P.; Weissman, J.S. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J. Cell Biol. 2009, 186, 323–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iqbal, J.; Dai, K.; Seimon, T.; Jungreis, R.; Oyadomari, M.; Kuriakose, G.; Ron, D.; Tabas, I.; Hussain, M.M. IRE1beta inhibits chylomicron production by selectively degrading MTP mRNA. Cell Metab. 2008, 7, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Pidoux, A.L.; Armstrong, J. Analysis of the BiP gene and identification of an ER retention signal in Schizosaccharomyces pombe. EMBO J. 1992, 11, 1583–1591. [Google Scholar] [PubMed]
- Guydosh, N.R.; Kimmig, P.; Walter, P.; Green, R. Regulated Ire1-dependent mRNA decay requires no-go mRNA degradation to maintain endoplasmic reticulum homeostasis in S. pombe. eLife 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Pineau, L.; Colas, J.; Dupont, S.; Beney, L.; Fleurat-Lessard, P.; Berjeaud, J.-M.; Bergès, T.; Ferreira, T. Lipid-induced ER stress: Synergistic effects of sterols and saturated fatty acids. Traffic 2009, 10, 673–690. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, T.; Nakayama, H.; Nagayoshi, Y.; Kakeya, H.; Kohno, S. Dissection of Ire1 functions reveals stress response mechanisms uniquely evolved in Candida glabrata. PLoS Pathog. 2013, 9, e1003160. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, T.; Kohno, S. ER stress response mechanisms in the pathogenic yeast Candida glabrata and their roles in virulence. Virulence 2014, 5, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, T.; Yamauchi, S.; Inamine, T.; Nagayoshi, Y.; Saijo, T.; Izumikawa, K.; Seki, M.; Kakeya, H.; Yamamoto, Y.; Yanagihara, K.; et al. Roles of calcineurin and Crz1 in antifungal susceptibility and virulence of Candida glabrata. Antimicrob. Agents Chemother. 2010, 54, 1639–1643. [Google Scholar] [CrossRef] [PubMed]
- Babour, A.; Bicknell, A.A.; Tourtellotte, J.; Niwa, M. A surveillance pathway monitors the fitness of the endoplasmic reticulum to control its inheritance. Cell 2010, 142, 256–269. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-L.; Konieczka, J.H.; Springer, D.J.; Bowen, S.E.; Zhang, J.; Silao, F.G.S.; Bungay, A.A.C.; Bigol, U.G.; Nicolas, M.G.; Abraham, S.N.; et al. Convergent Evolution of calcineurin pathway roles in thermotolerance and virulence in Candida glabrata. Genes Genome Genet. 2012, 2, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Askew, D.S. Endoplasmic reticulum stress and fungal pathogenesis converge. Virulence 2014, 5, 331–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishnan, K.; Askew, D.S. Endoplasmic reticulum stress and fungal pathogenesis. Fungal Biol. Rev. 2014, 28, 29–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, K.W.; So, Y.S.; Bahn, Y.S. Unique roles of the unfolded protein response pathway in fungal development and differentiation. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Cheon, S.A.; Jung, K.-W.; Chen, Y.-L.; Heitman, J.; Bahn, Y.-S.; Kang, H.A. Unique evolution of the UPR pathway with a novel bZIP transcription factor, Hxl1, for controlling pathogenicity of Cryptococcus neoformans. PLoS Pathog. 2011, 7, e1002177. [Google Scholar] [CrossRef] [PubMed]
- Cheon, S.A.; Jung, K.W.; Bahn, Y.S.; Kang, H.A. The Unfolded Protein Response (UPR) pathway in Cryptococcus. Virulence 2014, 5, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Liu, T.; Chen, L.; Li, W.; Liu, I.; Kronstad, J.W.; Seyfang, A.; Heitman, J. Role of an expanded inositol transporter repertoire in Cryptococcus neoformans sexual reproduction and virulence. mBio 2010, 1, e00084-10. [Google Scholar] [CrossRef] [PubMed]
- Glazier, V.E.; Kaur, J.N.; Brown, N.T.; Rivera, A.A.; Panepinto, J.C. Puf4 regulates both splicing and decay of HXL1 mRNA encoding the unfolded protein response transcription factor in Cryptococcus neoformans. Eukaryot. Cell 2015, 14, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.W.; Kang, H.A.; Bahn, Y.S. Essential roles of the Kar2/BiP molecular chaperone downstream of the UPR pathway in Cryptococcus neoformans. PLoS ONE 2013, 8, e58956. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Heitman, J. Function of Cryptococcus neoformans KAR7 (SEC66) in karyogamy during unisexual and opposite-sex mating. Eukaryot. Cell 2012, 11, 783–794. [Google Scholar] [CrossRef] [PubMed]
- Blankenship, J.R.; Fanning, S.; Hamaker, J.J.; Mitchell, A.P. An Extensive circuitry for cell wall regulation in Candida albicans. PLoS Pathog. 2010, 6, e1000752. [Google Scholar] [CrossRef] [PubMed]
- Azadmanesh, J.; Gowen, A.M.; Creger, P.E.; Schafer, N.D.; Blankenship, J.R. Filamentation involves two overlapping, but distinct, programs of filamentation in the pathogenic fungus Candida albicans. Genes Genome Genet. 2017, 7, 3797–3808. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-L.; Kauffman, S.; Reynolds, T.B. Candida albicans uses multiple mechanisms to acquire the essential metabolite inositol during infection. Infect. Immun. 2008, 76, 2793–2801. [Google Scholar] [CrossRef] [PubMed]
- Wimalasena, T.T.; Enjalbert, B.; Guillemette, T.; Plumridge, A.; Budge, S.; Yin, Z.; Brown, A.J.P.; Archer, D.B. Impact of the unfolded protein response upon genome-wide expression patterns, and the role of Hac1 in the polarized growth, of Candida albicans. Fungal Genet. Biol. 2008, 45, 1235–1247. [Google Scholar] [CrossRef] [PubMed]
- Thomas, E.; Sircaik, S.; Roman, E.; Brunel, J.M.; Johri, A.K.; Pla, J.; Panwar, S.L. The activity of RTA2, a downstream effector of the calcineurin pathway, is required during tunicamycin-induced ER stress response in Candida albicans. FEMS Yeast Res. 2015, 15, fov095. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Heitman, J.; Chen, Y.-L. Comparative analysis of calcineurin signaling between Candida dubliniensis and Candida albicans. Commun. Integr. Biol. 2012, 5, 122–126. [Google Scholar] [CrossRef] [PubMed]
- Morrow, M.W.; Janke, M.R.; Lund, K.; Morrison, E.P.; Paulson, B.A. The Candida albicans Kar2 protein is essential and functions during the translocation of proteins into the endoplasmic reticulum. Curr. Genet. 2011, 57, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, C.; Wang, A. Divergence and conservation of the major UPR branch IRE1-bZIP signaling pathway across eukaryotes. Sci. Rep. 2016, 6, 27362. [Google Scholar] [CrossRef] [PubMed]
- Montenegro-Montero, A.; Goity, A.; Larrondo, L.F. The bZIP transcription factor HAC-1 is involved in the unfolded protein response and is necessary for growth on cellulose in Neurospora crassa. PLoS ONE 2015, 10, e0131415. [Google Scholar] [CrossRef] [PubMed]









© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-Elvira, M.; Torres-Quiroz, F.; Escamilla-Ayala, A.; Domínguez-Martin, E.; Escalante, R.; Kawasaki, L.; Ongay-Larios, L.; Coria, R. The Unfolded Protein Response Pathway in the Yeast Kluyveromyces lactis. A Comparative View among Yeast Species. Cells 2018, 7, 106. https://doi.org/10.3390/cells7080106
Hernández-Elvira M, Torres-Quiroz F, Escamilla-Ayala A, Domínguez-Martin E, Escalante R, Kawasaki L, Ongay-Larios L, Coria R. The Unfolded Protein Response Pathway in the Yeast Kluyveromyces lactis. A Comparative View among Yeast Species. Cells. 2018; 7(8):106. https://doi.org/10.3390/cells7080106
Chicago/Turabian StyleHernández-Elvira, Mariana, Francisco Torres-Quiroz, Abril Escamilla-Ayala, Eunice Domínguez-Martin, Ricardo Escalante, Laura Kawasaki, Laura Ongay-Larios, and Roberto Coria. 2018. "The Unfolded Protein Response Pathway in the Yeast Kluyveromyces lactis. A Comparative View among Yeast Species" Cells 7, no. 8: 106. https://doi.org/10.3390/cells7080106
APA StyleHernández-Elvira, M., Torres-Quiroz, F., Escamilla-Ayala, A., Domínguez-Martin, E., Escalante, R., Kawasaki, L., Ongay-Larios, L., & Coria, R. (2018). The Unfolded Protein Response Pathway in the Yeast Kluyveromyces lactis. A Comparative View among Yeast Species. Cells, 7(8), 106. https://doi.org/10.3390/cells7080106