MicroRNAs at the Interface between Osteogenesis and Angiogenesis as Targets for Bone Regeneration
Abstract
:1. Introduction
2. Molecular Regulation of Bone Angiogenesis
3. The Role of MicroRNAs
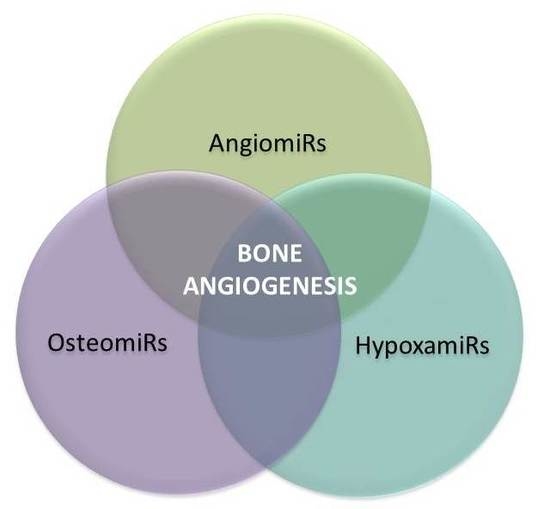
4. MicroRNAs in Bone Angiogenesis: OsteomiRs, AngiomiRs, and HypoxamiRs
5. Specific MicroRNAs Implicated in Angiogenic-Osteogenic Coupling
5.1. MiR-9
5.2. MiR-10a
5.3. MiR-10a/10b
5.4. MiR-20a
5.5. MiR-26a/b
5.6. MiR-29b
5.7. MiR-31
5.8. MiR-34a
5.9. MiR-92a
5.10. MiR-125b
5.11. MiR-135b
5.12. MiR-181a
5.13. MiR-195
5.14. MiR-200b
5.15. MiR-210
5.16. MiR-222
5.17. MiR-424
6. Outlook and Future Directions: MiRNAs in Therapeutic Applications
Author Contributions
Funding
Conflicts of Interest
References
- Crane, G.; Ishaug, S.; Mikos, A. Bone tissue engineering. Nat. Med. 1995, 1, 1322–1324. [Google Scholar] [CrossRef] [PubMed]
- Rouwkema, J.; Rivron, N.C.; Blitterswijk, C.A. Vascularization in tissue engineering. Trends Biotechnol. 2008, 26, 434–441. [Google Scholar] [CrossRef] [PubMed]
- Hertig, A. Angiogenesis in the early human chorion and the primary placenta of the macaque monkey. Contrib. Embryol. 1935, 25, 37–81. [Google Scholar]
- Chung, A.S.; Ferrara, N. Developmental and pathological angiogenesis. Annu. Rev. Cell Dev. Biol. 2011, 27, 563–584. [Google Scholar] [CrossRef]
- Lian, J.B.; Stein, G.S.; van Wijnen, A.J.; Stein, J.L.; Hassan, M.Q.; Gaur, T.; Zhang, Y. MicroRNA control of bone formation and homeostasis. Nat. Rev. Endocrinol. 2013, 8, 212–227. [Google Scholar] [CrossRef] [PubMed]
- Kronenberg HM Developmental regulation of the growth plate. Nature 2003, 423, 332–336. [CrossRef] [PubMed]
- Berendesen, A.; Olsen, B. Bone development. Bone 2015, 80, 14–18. [Google Scholar] [CrossRef] [Green Version]
- Olsen, B.; Reginato, A.; Wang, W. Bone development. Annu. Rev. Cell Dev. Biol. 2000, 16, 191–220. [Google Scholar] [CrossRef]
- Karsenty, G. The complexities of skeletal biology. Nature 2003, 423, 316–318. [Google Scholar] [CrossRef]
- Helms, J.; Schneider, R. Cranial skeletal biology. Nature 2003, 423, 326–331. [Google Scholar] [CrossRef]
- Maes, C.; Carmeliet, P.; Moermans, K.; Stockmans, I.; Smets, N. Impaired angiogenesis and endochondral bone formation in mice lacking the vascular endothelial growth factor isoforms VEGF 164 and VEGF 188. Mech Dev. 2002, 111, 61–73. [Google Scholar] [CrossRef]
- Gerber, H.; Ferrara, N. Angiogenesis and Bone Growth. TCM 2000, 10, 223–228. [Google Scholar] [CrossRef]
- Brandi, M.; Collin-Osdoby, P. Vascular biology and the skeleton. J. Bone Miner. Res. 2006, 21, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Sivaraj, K.K.; Adams, R.H. Blood vessel formation and function in bone. Development 2016, 143, 2706–2715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schipani, E.; Maes, C.; Carmeliet, G.; Semenza, G.L. Regulation of osteogenesis-angiogenesis coupling by HIFs and VEGF. J. Bone Miner. Res. 2009, 24, 1347–1353. [Google Scholar] [CrossRef] [PubMed]
- Schipani, E.; Wu, C.; Rankin, E.B.; Giaccia, A.J. Regulation of Bone Marrow Angiogenesis by Osteoblasts during Bone Development and Homeostasis. Front. Endocrinol. 2013, 4, 85. [Google Scholar] [CrossRef]
- Kusumbe, A.P.; Ramasamy, S.K.; Adams, R.H. Coupling of angiogenesis and osteogenesis by a specific vessel subtype in bone. Nature 2014, 507, 323–328. [Google Scholar] [CrossRef] [Green Version]
- Carmeliet, P.; Ferreira, V.; Breier, G.; Pollefeyt, S.; Kieckens, L.; Gertsenstein, M.; Fahrig, M.; Vandenhoeck, A.; Harpal, K.; Eerhardt, C.; et al. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature 1996, 380, 435–439. [Google Scholar] [CrossRef] [Green Version]
- Carulli, C.; Innocenti, M.; Brandi, M.L. Bone vascularization in normal and disease conditions. Front. Endocrinol. 2013, 4, 1–10. [Google Scholar] [CrossRef]
- Ding, W.-G.; Yan, W.; Wei, Z.-X.; Liu, J.-B. Difference in intraosseous blood vessel volume and number in osteoporotic model mice induced by spinal cord injury and sciatic nerve resection. J. Bone Miner. Metab. 2012, 30, 400–407. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, F.; Zhang, P.; Wang, H.; Qu, Z.; Jia, P.; Yao, Z.; Shen, G.; Li, G.; Zhao, G.; et al. Human type H vessels are a sensitive biomarker of bone mass. Cell Death Dis. 2017, 8, e2760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stegen, S.; Van Gastel, N.; Carmeliet, G. Bringing new life to damaged bone: The importance of angiogenesis in bone repair and regeneration. Bone 2014, 70, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.; Olsen, B.R. Osteoblast-derived VEGF regulates osteoblast differentiation and bone formation during bone repair. J. Clin. Invest. 2016, 126, 509–526. [Google Scholar] [CrossRef] [Green Version]
- Hu, K.; Olsen, B.R. The roles of vascular endothelial growth factor in bone repair and regeneration. Bone 2016, 91, 30–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maes, C.; Kobayashi, T.; Selig, M.; Torrekens, S.; Roth, S.; Mackem, S.; Carmeliet, G.; Kronenberg, H. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev. Cell 2010, 19, 329–344. [Google Scholar] [CrossRef] [PubMed]
- Maes, C.; Carmeliet, G.; Schipani, E. Hypoxia-driven pathways in bone development, regeneration and disease. Nat. Rev. Rheumatol. 2012, 8, 358–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Wan, C.; Deng, L.; Liu, X.; Cao, X.; Gilbert, S.R.; Bouxsein, M.L.; Faugere, M.; Guldberg, R.E.; Gerstenfeld, L.C.; et al. The hypoxia-inducible factor α pathway couples angiogenesis to osteogenesis during skeletal development. J. Clin. Invest. 2007, 117, 1616–1626. [Google Scholar] [CrossRef] [Green Version]
- Pugh, C.W.; Ratcliffe, P.J. Regulation of angiogenesis by hypoxia: Role of the HIF system. Nat. Med. 2003, 9, 677–684. [Google Scholar] [CrossRef]
- Ferrara, N.; Gerber, H.; Lecouter, J. The biology of VEGF and its receptors. Nat. Med. 2003, 9, 669–676. [Google Scholar] [CrossRef]
- Ornitz, D.; Marie, P. Fibroblast growth factor signaling in skeletal development and disease. Genes Dev. 2015, 29, 1463–1468. [Google Scholar] [CrossRef]
- Ramasamy, S.K.; Kusumbe, A.P.; Wang, L.; Adams, R.H. Endothelial Notch activity promotes angiogenesis and osteogenesis in bone. Nature 2014, 507, 376–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortega, N.; Behonick, D.; Werb, Z. Matrix remodeling during endochondral ossification. Trends Cell Biol. 2004, 14, 86–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleinheinz, J.; Stratmann, U.; Joos, U.; Wiesmann, H.-P. VEGF-Activated Angiogenesis During Bone Regeneration. J. Oral Maxillofac. Surg. 2005, 63, 1310–1316. [Google Scholar] [CrossRef] [PubMed]
- Maes, C.; Coenegrachts, L.; Stockmans, I.; Daci, E.; Luttun, A.; Petryk, A.; Gopalakrishnan, R.; Moermans, K.; Smets, N.; Verfaillie, C.M.; et al. Placental growth factor mediates mesenchymal cell development, cartilage turnover, and bone remodeling during fracture repair. J. Clin. Invest. 2006, 116, 16–18. [Google Scholar] [CrossRef]
- Kingsley, D. What do BMPs do in mammals? Clues from the mouse short-ear mutation. Trends Genet. 1994, 10, 16–21. [Google Scholar] [CrossRef]
- Hassan, M.Q.; Tye, C.E.; Stein, G.S.; Lian, J.B. Non-coding RNAs: Epigenetic regulators of bone development and homeostasis. Bone 2015, 81, 746–756. [Google Scholar] [CrossRef] [PubMed]
- Papaioannou, G.; Mirzamohammadi, F.; Kobayashi, T. MicroRNAs involved in bone formation. Cell Mol. Life Sci. 2014, 71, 4747–4761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papaioannou, G. miRNAs in Bone Development. Curr. Genom. 2015, 16, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V. MicroRNAs: Tiny regulators with great potential. Cell 2001, 107, 823–826. [Google Scholar] [CrossRef]
- Lau, N.C.; Lim, L.P.; Weinstein, E.G.; Bartel, D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 2001, 294, 858–862. [Google Scholar] [CrossRef]
- Bartel, D.P.; Lee, R.; Feinbaum, R. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function Genomics: The miRNA Genes. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, R.C.; Ambros, V. An extensive class of small RNAs in Caenorhabditis elegans. Science 2001, 294, 858–862. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, L.A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef] [PubMed]
- Hutvagner, G.; McLachlan, J.; Pasquinelli, A.E.; Balint, E.; Tuschl, T.; Zamore, P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Grishok, A.; Pasquinelli, A.E.; Conte, D.; Li, N.; Parrish, S.; Ha, I.; Baillie, D.L.; Fire, A.; Ruvkun, G.; Mello, C. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 2001, 106, 23–34. [Google Scholar] [CrossRef]
- Knight, S.W.; Bass, B.L. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 2001, 293, 2269–2271. [Google Scholar] [CrossRef]
- Yang, J.S.; Lai, E.C. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol. Cell 2011, 43, 892–903. [Google Scholar] [CrossRef]
- Lai, E.C. Micro RNAs are complementary to 3´UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 2002, 30, 363–364. [Google Scholar] [CrossRef]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef]
- Wang, Y.; Medvid, C.; Melton, R.; Jaenisch, R.; Blelloch, R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 2007, 39, 380–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Förstemann, K.; Tomari, Y.; Du, T.; Vagin, V.V.; Denli, A.M.; Bratu, D.P.; Klattenhoff, C.; Theurkauf, W.E.; Zamore, P.D. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double stranded RNA-binding domain protein. PLoS Biol. 2005, 3, e236. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Shen, X.J.; Zou, Q.; Wang, S.P.; Tang, S.M.; Zhang, G.Z. Biological functions of microRNAs: A review. J. Physiol. Biochem. 2011, 67, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Melton, C.; Judson, R.L.; Blelloch, R. Opposing micro-RNA families regulate self-renewal in mouse embryonic stem cells. Nature 2010, 463, 621–626. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, J.; Hipfner, D.R.; Stark, A.; Russell, R.B.; Cohen, S.M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 2003, 113, 25–36. [Google Scholar] [CrossRef]
- Hipfner, D.R.; Weigmann, K.; Cohen, S.M. The bantam gene regulates Drosophila growth. Genetics 2002, 161, 1527–1537. [Google Scholar] [PubMed]
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006, 3, 87–98. [Google Scholar] [CrossRef] [Green Version]
- Krützfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of microRNAs in vivo with ´antagomirs´. Nature 2005, 438, 685–689. [Google Scholar] [CrossRef]
- Miska, E.A. How microRNAs control cell division, differentiation and death. Curr. Opin. Genet. Dev. 2005, 15, 563–568. [Google Scholar] [CrossRef]
- Chen, C.Z.; Li, L.; Lodish, L.F.; Bartel, D. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004, 303, 83–86. [Google Scholar] [CrossRef]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez-Garcia, I.; Miska, E.A. MicroRNA functions in animal development and human disease. Development 2005, 132, 4653–4662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gennari, L.; Bianciardi, S.; Merlotti, D. MicroRNAs in bone diseases. Osteoporos. Int. 2017, 28, 1191–1213. [Google Scholar] [CrossRef] [PubMed]
- Clark, E.; Kalomoiris, S.; Nolta, J.; Fierro, F. Concise Review: MicroRNA Function in Multipotent Mesenchymal Stromal Cells. Stem Cells 2014, 32, 1074–1082. [Google Scholar] [CrossRef] [Green Version]
- Peng, S.; Gao, D.; Gao, C.; Wei, P.; Niu, M.; Shuai, C. MicroRNAs regulate signaling pathways in osteogenic differentiation of mesenchymal stem cells (Review). Mol. Med. Rep. 2016, 14, 623–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, S.; Deng, Y.; Gu, P.; Fan, X. MicroRNAs Regulate Bone Development and Regeneration. Int. J. Mol. Sci. 2015, 16, 8227–8253. [Google Scholar] [CrossRef] [Green Version]
- Ji, X.; Chen, X.; Yu, X. MicroRNAs in Osteoclastogenesis and Function: Potential Therapeutic Targets for Osteoporosis. Int. J. Mol. Sci. 2016, 17, 349. [Google Scholar] [CrossRef]
- Dong, S.; Yang, B.; Guo, H.; Kang, F. MicroRNAs regulate osteogenesis and chondrogenesis. Biochem. Biophys. Res. Commun. 2012, 418, 587–591. [Google Scholar] [CrossRef]
- Kiga, K.; Mimuro, H.; Suzuki, M.; Shinozaki-Ushiku, A.; Kobayashi, T.; Sanada, T.; Kim, M.; Ogawa, M.; Iwasaki, Y.W.; Kayo, H.; et al. Epigenetic silencing of miR-210 increases the proliferation of gastric epithelium during chronic Helicobacter pylori infection. Nat. Commun. 2014, 5, 4497. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Qiu, M.; Dou, C.; Cao, Z.; Dong, S. MicroRNAs in Bone Balance and Osteoporosis. Drug Dev. Res. 2015, 76, 235–245. [Google Scholar] [CrossRef]
- Nugent, M. MicroRNAs and Fracture Healing. Calcif. Tissue Int. 2017, 101, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Tian, B.O.; Qu, X.; Liu, F.; Tang, T.; Qin, A.N.; Zhu, Z.; Dai, K. MicroRNAs play a role in chondrogenesis and osteoarthritis (Review). Int. J. Mol. Med. 2014, 34, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Min, Z.; Zhang, R.; Yao, J.; Jiang, C.; Guo, Y.; Cong, F.; Wang, W.; Tian, J.; Zhong, N.; Sun, J.; et al. MicroRNAs associated with osteoarthritis differently expressed in bone matrix gelatin (BMG) rat model. Int. J. Clin. Exp. Med. 2015, 8, 1009–1017. [Google Scholar] [PubMed]
- Seeliger, C.; Balmayor, E.; van Griensven, M. miRNAs Related to Skeletal Diseases. Stem Cells Dev. 2016, 25, 1261–1281. [Google Scholar] [CrossRef] [PubMed]
- Anand, S.; Cheresh, D.A. Emerging Role of Micro-RNAs in the Regulation of Angiogenesis. Genes Cancer 2011, 2, 1134–1138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Olson, E.N. AngiomiRs—Key Regulators of Angiogenesis. Curr. Opin. Genet. Dev. 2009, 19, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Small, E.M.; Olson, E.N. Pervasive roles of microRNAs in cardiovascular biology. Nature 2011, 469, 336–342. [Google Scholar] [CrossRef] [Green Version]
- Weis, S.M.; Caheresh, D.A. Tumor angiogenesis: Molecular pathways and therapeutic targets. Nat. Med. 2011, 17, 1359–1370. [Google Scholar] [CrossRef]
- Salinas-Vera, Y.; Marchat, L.; Gallardo-Rincon, D.; Ruiz-Garcia, E.; Astudillo- De La Vega, H.; Echavarria-Zepeda, R.; Lopez-Camarillo, C. AngiomiRs: MicroRNAs driving angiogenesis in cancer (Review). Int. J. Mol. Med. 2018, 2018. [Google Scholar] [CrossRef]
- Suarez, Y.; Sessa, W.C. MicroRNAs As Novel Regulators of Angiogenesis. Circ. Res. 2009, 104, 442–454. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Yang, D.; Na, S.; Sandusky, G.; Zhang, Q.; Zhao, G. Dicer is required for embryonic angiogenesis during mouse development. J. Biol. Chem. 2005, 280, 9330–9335. [Google Scholar] [CrossRef] [PubMed]
- Albinsson, S.; Suarez, Y.; Skoura, A.; Offermann, S.; Miano, J.M.; Sessa, W.C. MicroRNAs are necessary for vascular smooth muscle growth, differentiation, and function. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 1118–1126. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Aurora, A.B.; Johnson, B.A.; Qi, X.; McAnnaly, J.; Hill, J.A.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell 2008, 15, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Suarez, Y.; Wang, C.; Manes, T.D.; Pober, J.S. Cutting edge: TNF-induced microRNAs regulate TNF-induced expression of E-selectin and intercellular adhesion molecule-1 on human endothelial cells: Feedback control of inflammation. J. Immunol. 2010, 184, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Suarez, Y.; Fernandez-Hernando, C.; Pober, J.; Sessa, W. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ. Res. 2007, 100, 1164–1173. [Google Scholar] [CrossRef] [PubMed]
- Greco, S.; Gaetano, C.; Martelli, F. HypoxamiR Regulation and Function in Ischemic Cardiovascular Diseases. Antioxid. Redox Signal. 2014, 21, 1202–1219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samanta, S.; Balasubramanian, S.; Rajasingh, S.; Patel, U.; Dhanasekaran, A.; Dawn, B.; Rajasingh, J. MicroRNA: A new therapeutic strategy for cardiovascular diseases. Trends Cardiovasc. Med. 2016, 26, 407–419. [Google Scholar] [CrossRef] [Green Version]
- Suarez, Y.; Fernandez-Hernando, C.; Yu, J.; Gerber, S.A.; Harrison, K.D.; Pober, J.S.; Iruela-Arispe, M.L.; Merkenschlager, M.; Sessa, W.C. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 14082–14087. [Google Scholar] [CrossRef] [Green Version]
- Würdinger, T.; Tannous, B.A.; Saydam, O.; Skog, J.; Grau, S.; Soutschek, J.; Weissleder, R.; Breakefield, X.O.; Krichevsky, A.M. miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell 2008, 14, 382–393. [Google Scholar] [CrossRef]
- Anand, S.; Cheresh, D.A. MicroRNA-mediated Regulation of the Angiogenic Switch. Curr. Opin. Hematol. 2011, 18, 171–176. [Google Scholar] [CrossRef]
- Nallamshetty, S.; Chan, S.Y.; Loscalzo, J. Hypoxia: A master regulator of microRNA biogenesis and activity. Free Radic. Biol. Med. 2013, 64, 20–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madanecki, P.; Kapoor, N.; Bebok, Z.; Ochocka, R.; Collawn, J.F.; Bartoszewski, R. Regulation of angiogenesis by hypoxia: The role of microRNA. Cell. Mol. Biol. Lett. 2013, 18, 47–57. [Google Scholar] [CrossRef] [PubMed]
- el Azzouzi, H.; Leptidis, S.; Doevendans, P.A.; De Windt, L.J. HypoxamiRs: Regulators of cardiac hypoxia and energy metabolism. Trends Endocrinol. Metab. 2015, 26, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Greco, S.; Martelli, F. MicroRNAs in Hypoxia Response. Antioxid. Redox Signal. 2014, 21, 1164–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collet, G.; Skrzypek, K.; Grillon, C.; Matejuk, A.; El Hafni-Rahbi, B.; Fayel, N.L.; Kieda, C. Hypoxia control to normalize pathologic angiogenesis: Potential role for endothelial precursor cells and miRNAs regulation. Vascul. Pharmacol. 2012, 56, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Bertero, T.; Rezzonico, R.; Pottier, N.; Mari, B. Impact of MicroRNAs in the Cellular Response to Hypoxia. Int. Rev. Cell Mol. Biol. 2017, 333, 91–158. [Google Scholar] [CrossRef] [PubMed]
- Hua, Z.; Lv, Q.; Ye, W.; Wong, A.C.-K.; Cai, G.; Gu, D.; Ji, Y.; Zhao, C.; Wang, J.; Yang, B.B.; et al. MiRNA-Directed Regulation of VEGF and Other Angiogenic Factors under Hypoxia. PloS ONE 2006, 1, e116. [Google Scholar] [CrossRef]
- Loscalzo, J. The cellular response to hypoxia: Tuning the system with microRNAs. J. Clin. Invest. 2010, 120, 3815–3817. [Google Scholar] [CrossRef]
- Devlin, C.; Greco, S.; Martelli, F.; Ivan, M. MiR-210: More than a silent player in hypoxia. IUBMB Life 2011, 63, 94–100. [Google Scholar] [CrossRef]
- Chan, S.; Loscalzo, J. MicroRNA-210: A unique and pleiotropic hypoxamir. Cell Cycle 2010, 9, 1072–1083. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, G.; Subramanian, I.V.; Adhikari, N.; Zhang, X.; Joshi, H.P.; Basi, D.; Chandrashekhar, Y.S.; Hall, J.L.; Roy, S.; Zeng, Y.; et al. Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF- α isoforms and promotes angiogenesis. J. Clin. Invest. 2010, 120, 4141–4154. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, A.; Yanagisawa, K.; Tanaka, M.; Cao, K.; Matsuyama, Y.; Goto, H.; Takahashi, T. Identification of hypoxia-inducible factor-1alpha as a novel target for miR-17-92 microRNA cluster. Cancer Res. 2008, 68, 5540–5545. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Tsai, M.; Hung, P.; Kao, S.; Liu, T.; Wu, K.; Chiou, S.; Lin, S.; Chang, K. miR31 ablates expression of the HIF regulatory factor FIH to activate the HIF pathway in head and neck carcinoma. Cancer Res. 2010, 70, 1635–1644. [Google Scholar] [CrossRef] [PubMed]
- Yuva-Aydemir, Y.; Simkin, A.; Gascon, E.; Gao, F.-B. MicroRNA-9: Functional evolution of a conserved small regulatory RNA. RNA Biol. 2011, 8, 557–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, R.; Kan, Q.; Sun, Y.; Wang, S.; Zhang, G.; Peng, T.; Jia, Y. MiR-9 promotes the neural differentiation of mouse bone marrow mesenchymal stem cells via targeting zinc finger protein 521. Neurosci. Lett. 2012, 515, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Lu, D.; Guo, H.; Miao, W.; Wu, G. MicroRNA-9 regulates osteoblast differentiation and angiogenesis via the AMPK signaling pathway. Mol. Cell Biochem. 2016, 411, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xu, H.; Kou, J.; Wang, Q.; Zheng, X.; Yu, T. MiR-9 promotes osteoblast differentiation of mesenchymal stem cells by inhibiting DKK1 gene expression. Mol. Biol. Rep. 2016, 43, 939–946. [Google Scholar] [CrossRef]
- Zhuang, G.; Wu, X.; Jiang, Z.; Kasman, I.; Yao, J.; Guan, Y.; Oeh, J.; Modrusan, Z.; Bais, C.; Sampath, D.; et al. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J. 2012, 31, 3513–3523. [Google Scholar] [CrossRef] [Green Version]
- Jones, S.; Watkins, G.; Le Good, N.; Roberts, S.; Murphy, C.; Brockbank, S.; Needham, M.; Read, S.; Newham, P. The identification of differentially expressed microRNA in osteoarthritic tissue that modulate the production of TNF-alpha and MMP13. Osteoarthr. Cartil. 2009, 17, 464–472. [Google Scholar] [CrossRef]
- Wang, S.; Tang, C.; Zhang, Q.; Chen, W. Reduced miR-9 and miR-181a expression down-regulates Bim concentration and promote osteoclasts survival. Int. J. Clin. Exp. Pathol. 2014, 7, 2209–2218. [Google Scholar]
- Li, J.; Zhang, Y.; Zhao, Q.; Wang, J.; He, X. MicroRNA-10a Influences Osteoblast Differentiation and Angiogenesis by Regulating ß-Catenin Expression. Cell. Physiol. Biochem. 2015, 37, 2194–2208. [Google Scholar] [CrossRef]
- Day, T.F.; Guo, X.; Garrett-Beal, L.; Yang, Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev. Cell 2005, 8, 739–750. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Deng, S.; Ma, Q.; Zhang, T.; Jia, C.; Zhuo, D.; Yang, F.; Wei, J.; Wang, L.; Dykxhoorn, D.M.; et al. MicroRNA-10A* and MicroRNA-21 Modulate Endothelial Progenitor Cell Senescence Via Suppressing High-Mobility Group A2. Circ. Res. 2013, 112, 152–164. [Google Scholar] [CrossRef]
- Hassel, D.; Cheng, P.; White, M.P.; Ivey, K.N.; Kroll, J.; Augustin, H.G.; Katus, H.A.; Stainier, D.Y.R.; Srivastava, D. MicroRNA-10 Regulates the Angiogenic Behavior of Zebrafish and Human Endothelial Cells by Promoting Vascular Endothelial Growth Factor Signaling. Circ. Res. 2012, 111, 1421–1433. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Ling, C.C.; Li, L.; Qin, Y.; Qi, J.; Liu, X.; You, B.; Shi, Y.; Zhang, J.; Xu, Q.J.H.; et al. MicroRNA-10a/10b represses a novel target gene mib1 to regulate angiogenesis. Cardiovasc. Res. 2016, 110, 140–150. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Fu, W.; He, M.; Xie, W.; Lv, Q.; Li, G.; Wang, H.; Lu, G.; Hu, X.; Jiang, S.; et al. MiRNA-20a promotes osteogenic differentiation of human mesenchymal stem cells by co-regulating BMP signaling. RNA Biol. 2011, 8, 829–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doebele, C.; Bonauer, A.; Fischer, A.; Scholz, A.; Ress, Y.; Urbich, C.; Hofmann, W.-K.; Zeiher, A.M.; Dimmeler, S. Members of the microRNA-17-92 cluster exhibit a cell-intrinsic antiangiogenic function in endothelial cells. Blood 2010, 115, 4944–4950. [Google Scholar] [CrossRef] [Green Version]
- Deng, H.-T.; Liu, H.-L.; Zhai, B.-B.; Zhang, K.; Xu, G.-C.; Peng, X.-M. Vascular endothelial growth factor suppresses TNFSF15 production in endothelial cells by stimulating miR-31 and miR-20a expression via activation of Akt and Erk signals. FEBS Open Bio. 2017, 7, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Luzi, E.; Marini, F.; Sala, S.C.; Tognarini, I.; Galli, G.; Brandi, M.L. Osteogenic Differentiation of Human Adipose Tissue–Derived Stem Cells Is Modulated by the miR-26a Targeting of the SMAD1 Transcription Factor. J. Bone Miner. Res. 2008, 23, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Luzi, E.; Marini, F.; Tognarini, I.; Galli, G.; Falchetti, A.; Brandi, M.L. The regulatory network menin-microRNA 26a as a possible target for RNA-based therapy of bone diseases. Nucleic. Acid Ther. 2012, 22, 103–108. [Google Scholar] [CrossRef]
- Su, X.; Liao, L.; Shuai, Y.; Jing, H.; Liu, S.; Zhou, H.; Liu, Y.; Jin, Y. MiR-26a functions oppositely in osteogenic differentiation of BMSCs and ADSCs depending on distinct activation and roles of Wnt and BMP signaling pathway. Cell Death Dis. 2015, 6, e1851. [Google Scholar] [CrossRef] [PubMed]
- Trompeter, H.-I.; Dreesen, J.; Hermann, E.; Iwaniuk, K.M.; Hafner, M.; Renwick, N.; Tuschl, T.; Wernet, P. MicroRNAs miR-26a, miR-26b, and miR-29b accelerate osteogenic differentiation of unrestricted somatic stem cells from human cord blood. BMC Genom. 2013, 14, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Icli, B.; Wara, A.K.M.; Moslehi, J.; Sun, X.; Plovie, E.; Cahill, M.; Marchini, J.F.; Schissler, A.; Padera, R.F.; Shi, J.; et al. MicroRNA-26a regulates pathological and physiological angiogenesis by targeting BMP/SMAD1 signaling. Circ. Res. 2013, 113, 1231–1241. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hassan, M.Q.; Jafferji, M.; Aqeilan, R.I.; Garzon, R.; Croce, C.M.; Van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. Biological Functions of miR-29b Contribute to Positive Regulation of Osteoblast Differentiation. J. Biol. Chem. 2009, 284, 15676–15684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossi, M.; Pitari, M.R.; Amodio, N.; Di Martino, T.M.; Conforti, F.; Leone, E.; Botta, C.; Paolino, F.M.; Giudice, T.D.E.L.; Iuliano, E.; et al. miR-29b Negatively Regulates Human Osteoclastic Cell Differentiation and Function: Implications for the Treatment of Multiple Myeloma-Related Bone Disease. J. Cell. Physiol. 2013, 228, 1506–1515. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Cai, H.-X.; Gao, S.; Yang, G.-L.; Deng, H.-T.; Xu, G.-C.; Han, J.; Zhang, Q.-Z.; Li, L.-Y. TNSF15 suppresses VEGF production in endothelial cells by stimulating miR-29b expression via activation of JNK-GATA3 Signals. Oncotarget 2016, 7, 69436–69449. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cai, B.; Shen, L.; Dong, Y.; Lu, Q.; Sun, S.; Liu, S.; Ma, S.; Ma, P.X.; Chen, J. MiRNA-29b suppresses tumor growth through simultaneously inhibiting angiogenesis and tumorigenesis by targeting Akt3. Cancer Lett. 2017, 397, 111–119. [Google Scholar] [CrossRef]
- Granchi, D.; Ochoa, G.; Leonardi, E.; Devescovi, V.; Baglìo, S.R.; Osaba, L.; Baldini, N.; Ciapetti, G. Gene expression patterns related to osteogenic differentiation of bone marrow-derived mesenchymal stem cells during ex vivo expansion. Tissue Eng. Part. C Methods 2010, 16, 511–523. [Google Scholar] [CrossRef]
- Baglìo, S.R.; Devescovi, V.; Granchi, D.; Baldini, N. MicroRNA expression profiling of human bone marrow mesenchymal stem cells during osteogenic differentiation reveals Osterix regulation by miR-31. Gene 2013, 527, 321–331. [Google Scholar] [CrossRef]
- Deng, Y.; Wu, S.; Zhou, H.; Bi, X.; Wang, Y.; Hu, Y.; Gu, P.; Fan, X. Effects of a miR-31, Runx2, and Satb2 regulatory loop on the osteogenic differentiation of bone mesenchymal stem cells. Stem Cells Dev. 2013, 22, 2278–2286. [Google Scholar] [CrossRef]
- Luo, J.; Lin, J.; Paranya, G.; Bischoff, J. Angiostatin Upregulates E-Selectin in Proliferating Endothelial Cells. Biochem. Biophys. Res. Commun. 1998, 911, 906–911. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Holmstrom, K.; Qiu, W.; Ditzel, N.; Shi, K.; Hokland, L.; Kassem, M. MicroRNA-34a Inhibits Osteoblast Differentiation and In Vivo Bone Formation of Human Stromal Stem Cells. Stem Cells 2014, 32, 902–912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, H.; Chen, H.; Huang, P.; Qi, J.; Qian, N.; Deng, L.; Guo, L. Glucocorticoids impair bone formation of bone marrow stromal stem cells by reciprocally regulating microRNA-34a-5p. Osteoporos. Int. 2016, 27, 1493–1505. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Jia, L.; Zheng, Y.; Jin, C.; Liu, Y.; Liu, H.; Zhou, Y. Mir-34a Promotes Osteogenic Differentiation of Human Adipose-Derived Stem Cells via the RBP2/NOTCH I/CYCLIN DI Coregulatory Network. Stem Cell Rep. 2016, 7, 236–248. [Google Scholar] [CrossRef] [PubMed]
- Engin, F.; Yao, Z.; Yang, T.; Zhou, G.; Bertin, T.; Jiang, M.M.; Chen, Y.; Wang, L.; Zheng, H.; Sutton, R.E.; et al. Dimorphic effects of Notch signaling in bone homeostasis. Nat. Med. 2008, 14, 299–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galindo, M.; Pratap, J.; Young, D.W.; Hovhannisyan, H.; Im, H.J.; Choi, J.Y.; Lian, J.B.; Stein, J.L.; Stein, G.S.; van Wijnen, A.J. The bone-specific expression of Runx2 oscillates during the cell cycle to support a G1-related antiproliferative function in osteoblasts. J. Biol. Chem. 2005, 280, 20274–20285. [Google Scholar] [CrossRef] [PubMed]
- Zha, X.; Sun, B.; Zhang, R.; Li, C.; Yan, Z.; Chen, J. Regulatory Effect of MicroRNA-34a on Osteogenesis and Angiogenesis in Glucocorticoid-Induced Osteonecrosis of the Femoral Head. J. Orthop. Res. 2018, 36, 417–424. [Google Scholar] [CrossRef]
- Zhao, T.; Li, J.; Chen, A.F. MicroRNA-34a induces endothelial progenitor cell senescence and impedes its angiogenesis via suppressing silent information regulator 1. Am. J. Endocrinol. Metab. 2010, 299, E110–E116. [Google Scholar] [CrossRef] [Green Version]
- Mattagajasingh, I.; Kim, C.; Naqvi, A.; Yamamori, T.; Hoffman, T.; Jung, S.; DeRicco, J.; Kasuno, K.; Irani, K. SIRT1 promotes endothelium-dependent vascular relaxation by activating endothelial nitric oxide synthase. Proc. Natl. Acad. Sci. USA 2007, 104, 14855–14860. [Google Scholar] [CrossRef] [Green Version]
- Kumar, B.; Yadav, A.; Lang, J.; Teknos, T.N.; Kumar, P. Dysregulation of MicroRNA-34a Expression in Head and Neck Squamous Cell Carcinoma Promotes Tumor Growth and Tumor Angiogenesis. PLoS ONE 2012, 7, e37601. [Google Scholar] [CrossRef]
- Chai, Z.T.; Kong, J.; Zhu, X.D.; Zhang, Y.Y.; Lu, L.; Zhou, J.M.; Wang, L.R.; Zhang, K.Z.; Zhang, Q.B.; Ao, J.Y.; et al. MicroRNA-26a Inhibits Angiogenesis by Down-Regulating VEGFA through the PIK3C2α/Akt/HIF-1α Pathway in Hepatocellular Carcinoma. PLoS ONE 2013, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Murata, K.; Ito, H.; Yoshitomi, H.; Yamamoto, K.; Fukuda, A.; Yoshikawa, J.; Furu, M.; Ishikawa, M.; Shibuya, H.; Matsuda, S. Inhibition of miR-92a enhances fracture healing via promoting angiogenesis in a model of stabilized fracture in young mice. J. Bone Miner. Res. 2014, 29, 316–326. [Google Scholar] [CrossRef] [PubMed]
- Kalinina, N.; Klink, G.; Glukhanyuk, E.; Lopatina, T.; Anastassia, E.; Akopyan, Z.; Tkachuk, V. miR-92a regulates angiogenic activity of adipose-derived mesenchymal stromal cells. Exp. Cell Res. 2015, 339, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Daniel, J.-M.; Penzkofer, D.; Teske, R.; Dutzmann, J.; Koch, A.; Bielenberg, W.; Bonauer, A.; Boon, R.A.; Fischer, A.; Bauersachs, J.; et al. Inhibition of miR-92a improves re-endothelialization and prevents neointima formation following vascular injury. Cardiovasc. Res. 2014, 103, 564–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goettsch, C.; Rauner, M.; Pacyna, N.; Hempel, U.; Bornstein, S.R.; Hofbauer, L.C. MiR-125b regulates calcification of vascular smooth muscle cells. Am. J. Pathol. 2011, 179, 1594–1600. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, Y.; Yagi, K.; Tokuzawa, Y.; Kanesaki-Yatsuka, Y.; Suda, T.; Katagiri, T.; Fukuda, T.; Maruyama, M.; Okuda, A.; Amemiya, T.; et al. miR-125b inhibits osteoblastic differentiation by down-regulation of cell proliferation. Biochem. Biophys. Res. Commun. 2008, 368, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Zhang, P.; Liang, P.; Huang, X. The expression of miR-125b regulates angiogenesis during the recovery of heat-denatured HUVECs. Burns 2015, 41, 803–811. [Google Scholar] [CrossRef]
- Huang, K.; Fu, J.; Zhou, W.; Li, W.; Dong, S.; Yu, S.; Hu, Z.; Wang, H.; Xie, Z. MicroRNA-125b regulates osteogenic differentiation of mesenchymal stem cells by targeting Cbfb in vitro. Biochimie 2014, 102, 47–55. [Google Scholar] [CrossRef]
- Xihong, L.U.; Min, D.; Honghui, H.E.; Dehui, Z.; Wei, Z. miR-125b regulates osteogenic differentiation of human bone marrow mesenchymal stem cells by targeting Smad4. J. Cent. South. Univ. (Med. Sci.) 2013, 38, 341–346. [Google Scholar] [CrossRef]
- Muramatsu, F.; Kidoya, H.; Naito, H.; Sakimoto, S.; Takakura, N. microRNA-125b inhibits tube formation of blood vessels through translational suppression of VE-cadherin. Oncogene 2013, 32, 414–421. [Google Scholar] [CrossRef]
- Schaap-Oziemlak, A.M.; Raymakers, R.A.; Bergevoet, S.M.; Gilissen, C.; Jansen, B.J.H.; Adema, G.J.; Kögler, G.; le Sage, C.; Agami, R.; van der Reijden, B.A.; et al. MicroRNA hsa-miR-135b Regulates Mineralization in Osteogenic Differentiation of Human Unrestricted Somatic Stem Cells. Stem Cells Dev. 2010, 19, 877–885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Umezu, T.; Tadokoro, H.; Azuma, K.; Yoshizawa, S.; Ohyashiki, K.; Ohyashiki, J.H. Exosomal miR-135b shed from hypoxic multiple myeloma cells enhances angiogenesis by targeting factor-inhibiting HIF-1. Blood 2014, 124, 3748–3757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, S.; Santini, G.C.; De Veirman, K. ; Broek, I.V.; Leleu, X.; De, A.; Van Camp, B.; Vanderkerken, K.; Van Riet, I. Upregulation of miR-135b Is Involved in the Impaired Osteogenic Differentiation of Mesenchymal Stem Cells Derived from Multiple Myeloma Patients. PLoS ONE 2013, 8, e79752. [Google Scholar] [CrossRef]
- Sumiyoshi, K.; Kubota, S.; Ohgawara, T.; Kawata, K.; Abd El Kader, T.; Nishida, T.; Ikeda, N.; Shimo, T.; Yamashiro, T.; Takigawa, M. Novel Role of miR-181a in Cartilage Metabolism. J. Cell. Biochem. 2013, 114, 2094–2100. [Google Scholar] [CrossRef] [PubMed]
- Gabler, J.; Ruetze, M.; Kynast, K.L.; Grossner, T.; Diederichs, S.; Richter, W. Stage-Specific miRs in Chondrocyte Maturation: Differentiation-Dependent and Hypertrophy-Related miR Clusters and the miR-181 Family. Tissue Eng. Part. A 2015, 21, 2840–2851. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Charbonneau, C.; Wei, L.; Chen, Q.; Terek, R.M. miR-181a Targets RGS16 to Promote Chondrosarcoma Growth, Angiogenesis, and Metastasis. Mol. Cancer Res. 2015, 13, 1347–1357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Wei, L.; Chen, Q.; Terek, R.M. MicroRNA Regulates Vascular Endothelial Growth Factor Expression in Chondrosarcoma Cells. Clin. Orthop. Relat. Res. 2015, 473, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Almeida, M.I.; Silva, A.M.; Vasconcelos, D.M.; Almeida, C.R.; Caires, H.; Pinto, M.T.; Calin, A.; Santos, S.G.; Barbosa, M.A. miR-195 in human primary mesenchymal stromal/stem cells regulates proliferation, osteogenesis and paracrine effect on angiogenesis. Oncotarget 2015, 7, 7–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; Zhao, N.; Li, S.; Fang, J.; Chen, M.; Yang, J.; Jia, W.; Yuan, Y.; Zhuang, S. MicroRNA-195 Suppresses Angiogenesis and Metastasis of Hepatocellular Carcinoma by Inhibiting the Expression of VEGF, VAV2, and CDC42. Hepatology 2013, 58, 642–653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Zeng, C.; Tu, M.; Jiang, W.; Dai, Z.; Hu, Y.; Deng, Z.; Xiao, W. MicroRNA-200b acts as a tumor suppressor in osteosarcoma via targeting ZEB1. Onco Targets Ther. 2016, 9, 3101–3111. [Google Scholar]
- Fan, X.; Teng, Y.; Ye, Z.; Zhou, Y.; Tan, W.-S. The effect of gap junction-mediated transfer of miR-200b on osteogenesis and angiogenesis in a co-culture of MSCs and HUVECs. J. Cell Sci. 2018, 131, jcs216135. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Yoon, S.; Jeong, Y.; Yoon, J.; Baek, K. Regulation of Vascular Endothelial Growth Factor Signaling by miR-200b. Mol. Cells 2011, 32, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, Y.; Tokuzawa, Y.; Ninomiya, Y.; Yagi, K.; Yatsuka-Kanesaki, Y.; Suda, T.; Fukuda, T.; Katagiri, T.; Kondoh, Y.; Amemiya, T.; et al. miR-210 promotes osteoblastic differentiation through inhibition of AcvR1b. FEBS Lett. 2009, 583, 2263–2268. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-D.; Cai, F.; Liu, L.; Zhang, Y.; Yang, A.-L. microRNA-210 is involved in the regulation of postmenopausal osteoporosis through promotion of VEGF expression and osteoblast differentiation. Biol. Chem. 2015, 396, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Fasanaro, P.; D’Alessandra, Y.; Di Stefano, V.; Melchionna, R.; Romani, S.; Pompilio, G.; Capogrossi, M.C.; Martelli, F. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand ephrin-A3. J. Biol. Chem. 2008, 283, 15878–15883. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Guo, D.; Yang, S.; Sun, H.; Wu, B.; Zhou, D. Inhibition of miR-222-3p activity promoted osteogenic differentiation of hBMSCs by regulating Smad5-RUNX2 signal axis. Biochem. Biophys. Res. Commun. 2016, 470, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Takigawa, S.; Chen, A.; Wan, Q.; Na, S.; Sudo, A.; Yokota, H.; Hamamura, K. Role of miR-222-3p in c-Src-Mediated Regulation of Osteoclastogenesis. Int. J. Mol. Sci. 2016, 17, 240. [Google Scholar] [CrossRef] [PubMed]
- Poliseno, L.; Tuccoli, A.; Mariani, L.; Evangelista, M.; Citti, L.; Woods, K.; Mercatanti, A.; Hammond, S.; Rainaldi, G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood 2006, 108, 3068–3071. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Yang, T.; Han, J.; Yan, K.; Qiu, X.; Zhou, Y.; Fan, Q.; Ma, B. MicroRNA Expression During Osteogenic Differentiation of Human Multipotent Mesenchymal Stromal Cells From Bone Marrow. J. Cell. Biochem. 2011, 112, 1844–1856. [Google Scholar] [CrossRef]
- Vimalraj, S.; Selvamurugan, N. MicroRNAs expression and their regulatory networks during mesenchymal stem cells differentiation toward osteoblasts. Int. J. Biol. Macromol. 2014, 66, 194–202. [Google Scholar] [CrossRef]
- Li, L.; Qi, Q.; Luo, J.; Huang, S.; Ling, Z.; Gao, M.; Zhou, Z.; Stiehler, M.; Zou, X. FOXO1-suppressed miR-424 regulates the proliferation and osteogenic differentiation of MSCs by targeting FGF2 under oxidative stress. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- de Pontual, L.; Yao, E.; Callier, P.; Faivre, L.; Drouin, V.; Cariou, S.; Van Haeringen, A.; Geneviève, D.; Goldenberg, A.; Oufadem, M.; Manouvrier, S.; Munnich, A.; et al. Germline deletion of the miR-17 ~ 92 cluster causes skeletal and growth defects in humans. Nat. Genet. 2011, 43, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- Penzkofer, D.; Bonauer, A.; Fischer, A.; Tups, A.; Brandes, R.P.; Zeiher, A.M.; Dimmeler, S. Phenotypic Characterization of miR-92a - /- Mice Reveals an Important Function of miR-92a in Skeletal Development. PLoS ONE 2014, 9, e101153. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Ma, J.; Chen, S.; Chen, X.; Yu, X. MicroRNA-17-92 cluster regulates osteoblast proliferation and differentiation. Endocrine 2014, 45, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Mao, G.; Wu, P.; Zhang, Z.; Zhang, Z.; Liao, W.; Li, Y.; Kang, Y. MicroRNA-92a-3p Regulates Aggrecanase-1 and Aggrecanase-2 Expression in Chondrogenesis and IL-1β- Induced Catabolism in Human Articular Chondrocytes. Cell. Physiol. Biochem. 2017, 44, 38–52. [Google Scholar] [CrossRef]
- Zhang, Z.; Kang, Y.; Zhang, Z.; Zhang, H.; Duan, X.; Liu, J.; Li, X.; Liao, W. Expression of microRNAs during chondrogenesis of human adipose-derived stem cells. Osteoarthr. Cartil. 2012, 20, 1638–1646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, G.; Zhang, Z.; Huang, Z.; Chen, W.; Huang, G.; Meng, F.; Zhang, Z.; Kang, Y. MicroRNA-92a-3p regulates the expression of cartilage-specific genes by directly targeting histone deacetylase 2 in chondrogenesis and degradation. Osteoarthr. Cartil. 2017, 25, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Bonauer, A.; Carmona, G.; Iwasaki, M.; Mione, M.; Koyanagi, M.; Fischer, A.; Burchfield, J.; Fox, H.; Doebele, C.; Ohtani, K.; et al. MicroRNA-92a Controls Angiogenesis and Functional Recovery of Ischemic Tissues in Mice. Science 2009, 324, 1710–1713. [Google Scholar] [CrossRef] [PubMed]
- Elmén, J.; Lindow, M.; Schütz, S.; Lawrence, M.; Petri, A.; Obad, S.; Lindholm, M.; Hedtjärn, M.; Hansen, H.; Berger, U.; et al. LNA-mediated microRNA silencing in non-human primates. Nature 2008, 452, 896–899. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhou, M.; Qin, G.; Weintraub, N.L.; Tang, Y. MiR-92a regulates viability and angiogenesis of endothelial cells under oxidative stress. Biochem. Biophys. Res. Commun. 2015, 446, 952–958. [Google Scholar] [CrossRef] [PubMed]
- Rippe, C.; Blimline, M.; Magerko, K.A.; Lawson, B.R.; LaRocca, T.; Donato, A.; Seals, D.R. MicroRNA Changes in Human Arterial Endothelial Cells with Senescence: Relation to Apoptosis, eNOS and Inflammation Catarina. Exp. Gerontol. 2012, 47, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Kim, H.K.; Chung, S.; Kim, K.S.; Dutta, A. Depletion of human micro-RNA miR-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation. J. Biol. Chem. 2005, 280, 16635–16641. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.-F.; Yang, G.-H.; Pan, X.-H.; Zhang, S.-J.; Zhao, C.; Qiu, B.-S.; Gu, H.-F.; Hong, J.-F.; Cao, L.; Chen, Y.; et al. Altered MicroRNA Expression Profile in Exosomes during Osteogenic Differentiation of Human Bone Marrow- Derived Mesenchymal Stem Cells. PLoS ONE 2014, 9, e114627. [Google Scholar] [CrossRef] [PubMed]
- Fan, G. Hypoxic exosomes promote angiogenesis Platelets: Balancing the septic triad. Blood 2014, 124, 3669–3670. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Sun, Z.-J.; Bian, Y.; Kulkarni, A.B. MicroRNA-135b acts as a tumor promoter by targeting the hypoxia-inducible factor pathway in genetically defined mouse model of head and neck squamous cell carcinoma. Cancer Lett. 2013, 331, 230–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kazenwadel, J.; Michael, M.Z.; Harvey, N.L. Prox1 expression is negatively regulated by miR-181 in endothelial cells. Blood 2010, 116, 2395–2401. [Google Scholar] [CrossRef] [Green Version]
- Naguibneva, I.; Ameyar-Zazoua, M.; Polesskaya, A.; Ait-Si-Ali, S.; Groisman, R.; Souidi, M.; Cuvellier, S.; Harel-Bellan, A. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat. Cell Biol. 2006, 8, 278–284. [Google Scholar] [CrossRef]
- Akiyama, T.; Bouillet, P.; Miyazaki, T.; Kadono, Y.; Chikuda, H.; Chung, U.; Fukuda, A.; Hikita, A.; Seto, H.; Okada, T.; et al. Regulation of osteoclast apoptosis by ubiquitination of proapoptotic BH3-only Bcl-2 family member Bim. EMBO J. 2003, 22, 6653–6664. [Google Scholar] [CrossRef]
- Palmieri, A.; Pezzetti, F.; Brunelli, G.; Zollino, I.; Scapoli, L.; Martinelli, M.; Arlotti, M.; Carinci, F. Differences in osteoblast miRNA induced by cell binding domain of collagen and silicate-based synthetic bone. J. Biomed. Sci. 2007, 14, 777–782. [Google Scholar] [CrossRef]
- Christoffersen, N.R.; Silahtaroglu, A.; Ørom, U.L.F.A.; Kauppinen, S.; Lund, A.H. miR-200b mediates post-transcriptional repression of ZFHX1B. RNA 2007, 13, 1172–1178. [Google Scholar] [CrossRef]
- Baglio, S.R.; Rooijers, K.; Koppers-lalic, D.; Verweij, F.J.; Lanzón, M.P.; Zini, N.; Naaijkens, B.; Perut, F.; Niessen, H.W.M.; Baldini, N.; et al. Human bone marrow- and adipose- mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res. Ther. 2015, 6, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Zong, L.; Zhu, Y.; Liang, R.; Zhao, H.-B. Gap junction mediated miRNA intercellular transfer and gene regulation: A novel mechanism for intercellular genetic communication. Sci. Rep. 2016, 6, 19884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.-T.; Chen, H.-T.; Tsou, H.-K.; Tan, T.-W.; Fong, Y.-C.; Chen, P.-C.; Yang, W.-H.; Wang, S.-W.; Chen, J.-C.; Tang, C.-H. CCL5 promotes VEGF-dependent angiogenesis by downregulating miR-200b through PI3K/Akt signaling pathway in human chondrosarcoma cells. Oncotarget 2014, 5, 10718–10731. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.C.; Khanna, S.; Roy, S.; Sen, C.K. miR-200b targets Ets-1 and is down-regulated by hypoxia to induce angiogenic response of endothelial cells. J. Biol. Chem. 2011, 286, 2047–2056. [Google Scholar] [CrossRef] [PubMed]
- Lou, Y.-L.; Guo, F.; Liu, F.; Gao, F.-L.; Zhang, P.-Q.; Niu, X.; Guo, S.-C.; Yin, J.-H.; Wang, Y.; Deng, Z.-F. miR-210 activates notch signaling pathway in angiogenesis induced by cerebral ischemia. Mol. Cell Biochem. 2012, 370, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Fasanaro, P.; Greco, S.; Lorenzi, M.; Pescatori, M.; Brioschi, M.; Kulshreshta, R.; Banfi, C.; Stubbs, A.; Calin, G.A.; Ivan, M.; et al. An integrated approach for experimental target identification of hypoxia-induced miR-210. J. Biol. Chem. 2009, 284, 35134–35143. [Google Scholar] [CrossRef] [PubMed]
- Ivan, M.; Huang, X. miR-210: Fine-Tuning the Hypoxic Response. Adv. Exp. Med. Biol. 2014, 772, 205–227. [Google Scholar] [CrossRef]
- Ivan, M.; Harris, A.L.; Martelli, F.; Kulshreshtha, R. Hypoxia response and microRNAs: No longer two separate worlds. J. Cell Mol. Med. 2008, 12, 1426–1431. [Google Scholar] [CrossRef]
- Kuijper, S.; Turner, C.J.; Adams, R.H. Regulation of angiogenesis by Eph-ephrin interactions. Trends Cardiovasc. Med. 2007, 17, 145–151. [Google Scholar] [CrossRef]
- Pandey, A.; Shao, H.; Marks, R.M.; Polverini, P.J.; Dixit, V.M. Role of B61, the ligand for the Eck receptor tyrosine kinase, in TNF-alpha-induced angiogenesis. Science 1995, 268, 567–569. [Google Scholar] [CrossRef]
- Matsui, J.; Wakabayashi, T.; Asada, M.; Yoshimatsu, K. Stem cell factor/c-kit signaling promotes the survival, migration, and capillary tube formation of human umbilical vein endothelial cells. J. Biol. Chem. 2004, 279, 18600–18607. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Kleinheinz, J. Angiogenesis—The Key to Regeneration. In Tissue Engineering and Regenerative Medicine; Andrades, J.A., Ed.; InTechOpen: London, UK, 2013; pp. 453–473. [Google Scholar]
- Kanczler, J.M.; Oreffo, R.O.C. Osteogenesis and angiogenesis: The potential for engineering bone. Eur. Cells Mater. 2008, 15, 100–114. [Google Scholar] [CrossRef]
- Hou, H.; Zhang, X.; Tang, T.; Dai, K.; Ge, R. Enhancement of bone formation by genetically-engineered bone marrow stromal cells expressing BMP-2, VEGF and angiopoietin-1. Biotechnol. Lett. 2009, 31, 1183–1189. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Qiu, T.; Wu, X.; Wan, C.; Shi, W.; Wang, Y.; Chen, J.; Wan, M.; Clemens, T.L.; Cao, X. Sustained BMP Signaling in Osteoblasts Stimulates Bone Formation by Promoting Angiogenesis and Osteoblast Differentiation. J. Bone Miner. Res. 2009, 24, 1224–1233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Z.; Wang, K. Effects of recombinant adeno-associated viral vectors on angiopoiesis and osteogenesis in cultured rabbit bone marrow stem cells via co-expressing hVEGF and hBMP genes: A preliminary study in vitro. Tissue Cell 2010, 42, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Hon, L.S.; Zhang, Z. The roles of binding site arrangement and combinatorial targeting in microRNA repression of gene expression. Genome Biol. 2007, 8, R166. [Google Scholar] [CrossRef]
- Grimson, A.; Farh, K.K.; Johnston, W.K.; Garrett-Engele, P.; Lim, L.P.; Bartel, D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell 2007, 27, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Betel, D.; Wilson, M.; Gabow, A.; Marks, D.S.; Sander, C. The microRNA.org resource: Targets and expression. Nucleic Acids Res. 2008, 36, D149–D153. [Google Scholar] [CrossRef]
- Krek, A.; Grün, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; da Piedade, I.; Gunsalus, K.C.; Stoffel, M.; et al. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef]
- Brennecke, J.; Stark, A.; Russell, R.; Cohen, S. Principles of microRNA-target recognition. PLoS Biol. 2005, 3, e85. [Google Scholar] [CrossRef]
- Tsang, J.; Zhu, J.; van Oudenaarden, A. MicroRNAmediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell 2007, 26, 753–767. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.; Burge, C.; Bartel, D. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xie, H.; Liu, W.; Hu, R.; Huang, B.; Tan, Y.; Xu, K.; Sheng, Z.; Zhou, H.; Wu, X.; et al. A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J. Clin. Invest. 2009, 119, 3666–3677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mariner, P.; Johannesen, E.; Anseth, K. Manipulation of miRNA activity accelerates osteogenic differentiation of hMSCs in engineered 3D scaffolds. J. Tissue Eng. Regen Med. 2012, 6, 314–324. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.Y.; Li, N.; Lin, S.; Wang, B.; Lan, H.Y.; Li, G. miRNA-29b improves bone healing in mouse fracture model. Mol. Cell. Endocrinol. 2016, 430, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fan, L.; Liu, S.; Liu, W.; Zhang, H.; Zhou, T.; Wu, D.; Yang, P.; Shen, L.; Chen, J.; et al. The promotion of bone regeneration through positive regulation of angiogenic-osteogenic coupling using microRNA-26a. Biomaterials 2013, 34, 5048–5058. [Google Scholar] [CrossRef] [PubMed]
- Yoshizuka, M.; Nakasa, T.; Kawanishi, Y.; Hachisuka, S.; Furuta, T.; Miyaki, S.; Adachi, N.; Ochi, M. Inhibition of microRNA-222 expression accelerates bone healing with enhancement of osteogenesis, chondrogenesis, and angiogenesis in a rat refractory fracture model. J. Orthop. Sci. 2016, 21, 852–858. [Google Scholar] [CrossRef] [PubMed]
- Ørom, U.A.; Kauppinen, S.; Lund, A.H. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene 2006, 372, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Adams, B.D.; Parsons, C.; Walker, L.; Zhang, W.C.; Slack, F.J. Targeting noncoding RNAs in disease. J. Clin. Investig. 2017, 127, 761–771. [Google Scholar] [CrossRef] [Green Version]
- Simonson, B.; Das, S. MicroRNA Therapeutics: The Next Magic Bullet? Mini Rev. Med. Chem. 2016, 15, 467–474. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs-microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef] [PubMed]
- López-Camarillo, C.; Marchat, L.A.; Aréchaga-Ocampo, E.; Azuara-Liceaga, E.; Pérez-Plasencia, C.; Fuentes-Mera, L.; Fonseca-Sánchez, M.A.; Flores-Pérez, A. Functional Roles of microRNAs in Cancer: microRNomes and oncomiRs Connection; Oncogenomi.; In Tech Open Science: London, UK, 2013. [Google Scholar]
- Senanayake, U.; Das, S.; Vesely, P.; Alzoughbi, W.; Fröhlich, L.F.; Chowdhury, P.; Leuschner, I.; Hoefler, G.; Guertl, B. miR-192, miR-194, miR-215, miR-200c and miR-141 are downregulated and their common target ACVR2B is strongly expressed in renal childhood neoplasms. Carcinogenesis 2012, 33, 1014–1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| MicroRNAs | Targets 1 | Regulatory Role | Effects | Study Models | Ref. |
|---|---|---|---|---|---|
| MiR-9 | VEGF, VE-CAD (CD144) | AMPK signaling pathway | Enhanced osteogenic diff. & mineral.; increased angiogenesis | MC3T3-E1 | [106] |
| DKK1 | COL1, OCN, BSP; ALP activity | OB diff. & mineralization | C2C12 cells | [107] | |
| SOCS5 | JAK-STAT signaling pathway | Promotion of EC migration & angiogenesis | Primary microvascular ECs, HUVECs | [108] | |
| Cbl | Bim ubiquitination, apoptosis | Promotion of OC survival | OC, OC precursor cells (RAW264.7) | [110] | |
| MiR-10a | β-catenin, LEF1; VEGF, VE-CAD (CD144), cyclin D1, MMP2 | Wnt signaling; angiogenesis-related gene expression | Inhibition of osteogenic diff. & blood vessel formation | MC3T3-E1 MUVECS | [111] |
| HMGA2 | β–galactosidase expr; p16Ink4a/p19Arf expression | EPC senescence & angiogenesis; self-renewal potential | lin−BM-MSCs | [113] | |
| MiR-10a/10b | MIB1 | Notch signaling | Regulating blood vessel outgrowth/tip cell behavior | HUVECs | [115] |
| MiR-20a | BMP2, BMP4, RUNX2 | Effects BMP/RUNX2 signaling positively; blocks OB inhibitors & PPARγ | Enhances osteogenic differentiation; suppresses adipogenesis | hBM-MSC | [116] |
| JAK1; p21, S1P receptor EDG | Downregulation of proangiogenic JAK 1 & cell cycle inhibitors | Inhibits EC sprout formation | HUVECs | [117] | |
| TNFSF15 | VEGF-AKT/ERK –miR20a/31 signaling | Stimulation of angiogenesis | HUVECs | [118] | |
| MiR-26a | VEGF, ANG1, RUNX2, BMP2 OCN, ALP; GSK3β | WNT signaling activation | Enhanced angiogenesis & bone regeneration | Primary hBM-MSC, MC3T3-E1 | [76,98] |
| VEGF | PIK3C2α/AKT/HIF-α/VEGFA pathway | Inhibition of angiogenesis; | HUVECs | [141] | |
| SMAD1 | BMP signaling inhibition | OB differentiation | hADSCs | [119] | |
| SMAD1 | BMP signaling | Inhibits EC growth, proliferation, migration; regulates early angiogenesis | HUVECs | [123] | |
| MiR-29b | TGF-β3, HDAC4, ACTVR2A, CTNNBIP1, DUSP2; COL1A1, 5A3, 4A2 | Silences neg. osteogenic regulators suppresses ECM protein synthesis | Promotes osteoblastogenesis at multiple stages | MC3T3 pre-OB | [124] |
| c-FOS | Reduced TRAP expr., lacunae generation, collagen degradation | Neg. regulator of human OC differentiation and activity | OC (CD14 +) | [125] | |
| TNFSF15 | TNFSF15-enhanced JNK-GATA3 signal. & VEGF inhibition | Suppression of VEGF secretion | Mouse EC line bEnd.3 | [126] | |
| AKT3 | Inhibition of tumor vascularization via VEGF & cancer cell activity via c-MYC | Anti-angiogenic and anti-tumorigenic role | HUVECs, Breast cancer cells | [127] | |
| MiR-31 | OSX | Downregulation of OSX | Influences osteogenic differentiation | hMSC; Osteosarcoma cell | [129] |
| Satb2 protein | Inhibition by RUNX2; Upregulation of Satb2 protein & osteogenic TF | Induces BM-MSC osteogenic differentiation | hBM-MSC | [130] | |
| E-selectin | Regulation of E-selectin expression | Inhibition of angiostatin-induced angiogenesis; TNF-mediated induction of endothelial adhesion | HUVECs | [84] | |
| TNFSF15 | VEGF-AKT/ERK –miR20a/31 signaling | Stimulation of angiogenesis | HUVECs | [118] | |
| MiR-34a | Jagged1 | Regulation of cell cycle regulator & proliferation proteins & Jagged1 | Inhibition of osteoblast differentiation | hMSC; mouse heterotopic bone formation model | [132] |
| JAGGED1 | Activation of Notch signaling | Induction of glucocorticoid-mediated osteogenic differentiation | hMSC | [133] | |
| RBP2 | Promotes mineral, ALP activity & RUNX2 expression; downreg. NOTCH1 & Cyclin D1 expr. | Promotion of osteogenic differentiation; enhanced heterotopic bone formation | hADSCs; mouse heterotopic bone formation model | [134] | |
| VEGF | Inhibitory effects of dexamethasone on EC viability & VEGF | Decreased blood vessel development | Rat Glucocorticoid- induced osteonecrosis | [137] | |
| SIRT1 | Increased SIRT1 expr. & FOXO1 acetylation regulating vascular EC homeostasis | Inhibition of EPC-mediated angiogenesis | Rat EPC | [138] | |
| E2F3a, survivin | Interference with VEGF secretion, EC proliferation & migration | Dysregulated tumor angiogenesis | HNSCC tumors & cells | [140] | |
| MiR-92a | ? | ? | Enhanced fracture healing & inhib. of neovascularization | Mice with femoral fracture | [142] |
| HGF, ANGPT1 | ITGA5, MEK4 | Inhibition of tube formation by HUVECs | hADSCs | [143] | |
| ? | integrin a5, sirtuin1, eNOS | Attenuates neointimal lesion by accelerating re-endothelialization | MiR-92a knockout mice | [144] | |
| MiR-125b | OSX | RUNX2, a-SMC, ALP, matrix mineralization | Calcification of vascular smooth muscle cells | HCASMCs | [145] |
| ErbB2 | ? | Inhibits OB diff by downreg. of cell proliferation | ST2 cells (mMSCs) | [146] | |
| VEGF, ERBB2 | Regulation of angiogenesis during wound healing | HUVECS | [147] | ||
| Cbf-beta | ALP, OCN, OPN | Inhibition of osteogenic differentiation | C3H10T1/2 | [148] | |
| SMAD4 | ALP, RUNX2 | Downregulation of osteogenic differentiation | hMSCs | [149] | |
| VE-Cadherin | Inhibition of blood vessel (tube) formation | HUVECs | [150] | ||
| MiR-135b | ? | ? | OB differentiation | hBM-SCs | [151] |
| HIF-1 | ? | Enhanced endothelial tube formation | Human MM cells; HUVECs | [152] | |
| SMAD5 | ? | Impaired osteogenic differentiation | hMSCs | [153] | |
| MiR-181a | ? | CCN1, aggrecan | Maintaining homeostasis of chondrocytes | Human HCS-2/8 cells | [154] |
| COL10A1 | Chondrocyte differentiation | hMSC | [155] | ||
| RGS16 | CXCR4 signaling; VEGF, MMP1 | Angiogenesis & metastasis in chondrosarcoma | Xenograft mice; JJ chondrosarc. cells | [156] | |
| ? | VEGF expression | Chondrosarcoma-associated angiogenesis | JJ chondrosarc. cell line | [157] | |
| Cbl | Bim ubiquitination, apoptosis | promote OC survival | OC, OC precursor cells (RAW264.7) | [110] | |
| MiR-195 | ? | VEGF | Osteogenic diff. & proliferation; control of angiogenesis | hMSC(MC3T3) chick chorio-allantoic membrane | [158] |
| ? | VEGF, VAV2 CDC42 | HCC-associated angiogenesis & metastasis; migration & capillary tube form. of ECs | QGY-7703, MHCC-97H HCC cells; HUVECs | [159] | |
| MiR-200b | ZEB1 | ZEB1-TF target genes | Inhibits proliferation, migration & invasion of osteosarcoma cells | OsteosarcomaU2OS, Saos2, HOS, MG63 | [160] |
| VEGF-A; ZEB2, ETS1, KDR,GATA2 | Decreases VEGF-A expression & TF-target genes | Inhibition of VEGF-A induced osteogenesis; Inhibition of TF-activated angiogenesis | Rat BM-MSC & HUVEC coculture | [161] | |
| VEGF, FLT-1, and KDR | VEGF-induced phosph. of ERK1/2 | Inhibition of angiogenesis; red. capillary formation | A549 cells, HUVECs | [162] | |
| MiR-210 | AcvR1b | Inhib. of TGFb/activin signaling | Promotes OB differentiation | ST2 stromal cells | [163] |
| VEGF | PPARgamma, ALP, OSX | Promoteion of OB diff., inhibition of adipocyte diff. | hBM-SCs, 17β-estradiol (E2)treated OB | [164] | |
| EFNA3 | VEGF-expression mediated angiogenesis | EC survival, diff., migration; stim. of tubulogen. & chemotaxis | HUVECs | [165] | |
| MiR-222 | SMAD 1, 5, 8 protein & phosphoryl. | Decreased SMAD5-RUNX2 signaling & OSX, ALP, and OC levels & mineral. | Neg. regulator of osteogenic differentiation | hBM-SC | [166] |
| c-Src, Dcstamp | RANKL-induced expression of TRAP & cathepsin K | Inhibitory regulator of c-Src-mediated osteoclastogenesis | RAW264.7 pre-OC cells | [167] | |
| c-KIT | Suppression of tube formation, wound healing, cell migration via SCF | Inhibitory regulation of in vitro angiogenesis | HUVEC | [168] | |
| MiR-424 | RUNX, CBFβ, BMP | Osteogenic diff. of hMSCs | Bone formation | hMSCs | [169] |
| MAPK, WNT & insulin signal. | OB differentiation of hMSCs | Bone formation | hMSCs | [170] | |
| FGF-2; via FOXO1 | Decrease of ALP, mineralization & osteog. markers | Enhances proliferation & osteogenic differentiation of hMSCs | Pigs, cellular oxidative stress model | [171] | |
| CUL2; via RUNX-1→ C/EBPα→ PU.1 | Stabilization of HIF-1α | Regulation of Angiogenesis | ECs, ischemic tissues | [101] |
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fröhlich, L.F. MicroRNAs at the Interface between Osteogenesis and Angiogenesis as Targets for Bone Regeneration. Cells 2019, 8, 121. https://doi.org/10.3390/cells8020121
Fröhlich LF. MicroRNAs at the Interface between Osteogenesis and Angiogenesis as Targets for Bone Regeneration. Cells. 2019; 8(2):121. https://doi.org/10.3390/cells8020121
Chicago/Turabian StyleFröhlich, Leopold F. 2019. "MicroRNAs at the Interface between Osteogenesis and Angiogenesis as Targets for Bone Regeneration" Cells 8, no. 2: 121. https://doi.org/10.3390/cells8020121
APA StyleFröhlich, L. F. (2019). MicroRNAs at the Interface between Osteogenesis and Angiogenesis as Targets for Bone Regeneration. Cells, 8(2), 121. https://doi.org/10.3390/cells8020121