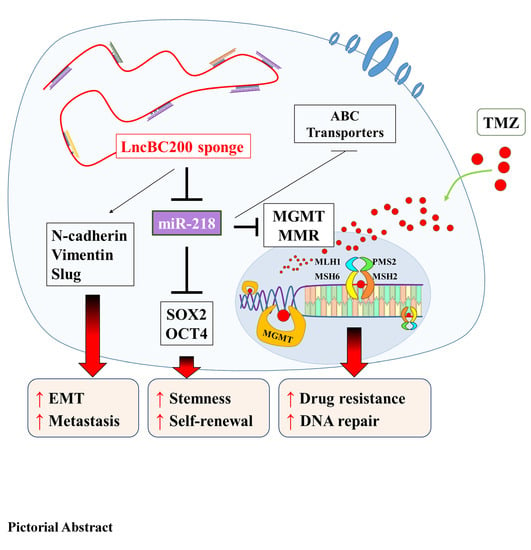
Targeting BC200/miR218-5p Signaling Axis for Overcoming Temozolomide Resistance and Suppressing Glioma Stemness
Abstract
:1. Background
2. Materials and Methods
2.1. Patients and Tumor Samples
2.2. Cell Lines and Cell Culture
2.3. Sulforhodamine B (SRB) Viability Assay
2.4. Cell Proliferation Assay
2.5. Vector Construction and Infection
2.6. Immunohistochemistry
2.7. Western Blot and RT-qPCR
2.8. Colony Formation Assay
2.9. Wound Healing Migration Assay
2.10. Matrigel Invasion Assay
2.11. Sphere Formation Assay
2.12. Flow Cytometry
2.13. In Vivo Studies
2.14. Statistical Analysis
3. Results
3.1. Upregulation of BC200 RNA Expression in GB Patients
3.2. High BC200 RNA Expression Associated with TMZ Resistance of Stem-Cell-Like Population in GB Cells
3.3. BC200 RNA Silencing Inhibits the Proliferation, Migration, Invasion, and Self-Renewal Ability of GB Cells
3.4. BC200 RNA Overexpression Enhances Aggresivenesss Behevior and Self-Renewal Ability of GB Cells
3.5. BC200 RNA Expression Associated with TMZ Resistance and miR-218-5p Expression
3.6. miR-218-5p Modulates Stem Cell Characteristics and TMZ Resistance
3.7. BC200 RNA Inactivity Sensitized GB Cells to TMZ Combined Therapy In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
Ethics approval and consent to participate
Availability of data and materials
References
- Hanif, F.; Muzaffar, K.; Perveen, K.; Malhi, S.M.; Simjee, S.U. Glioblastoma Multiforme: A Review of its Epidemiology and Pathogenesis through Clinical Presentation and Treatment. Asian Pac. J. Cancer Prev. 2017, 18, 3–9. [Google Scholar]
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Burger, P.C.; Jouvet, A.; Scheithauer, B.W.; Kleihues, P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007, 114, 97–109. [Google Scholar] [CrossRef] [Green Version]
- Jovcevska, I.; Kocevar, N.; Komel, R. Glioma and glioblastoma—how much do we (not) know? Mol. Clin. Oncol. 2013, 1, 935–941. [Google Scholar] [CrossRef] [Green Version]
- Lara-Velazquez, M.; Al-Kharboosh, R.; Jeanneret, S.; Vazquez-Ramos, C.; Mahato, D.; Tavanaiepour, D.; Rahmathulla, G.; Quinones-Hinojosa, A. Advances in Brain Tumor Surgery for Glioblastoma in Adults. Brain Sci. 2017, 7, 166. [Google Scholar] [CrossRef] [Green Version]
- Uszczynska-Ratajczak, B.; Lagarde, J.; Frankish, A.; Guigó, R.; Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 2018, 19, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Sun, S.; Pu, J.K.S.; Tsang, A.C.O.; Lee, D.; Man, V.O.Y.; Lui, W.M.; Wong, T.S.; Leung, G.K.K. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol. Dis. 2012, 48, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Zhang, Q.C.; Georgiev, P.; Ilik, I.; Akhtar, A.; Chang, H.Y. Rapid evolutionary turnover underlies conserved lncRNA-genome interactions. Genes Dev. 2016, 30, 191–207. [Google Scholar] [CrossRef] [Green Version]
- Cai, T.; Liu, Y.; Xiao, J. Long noncoding RNA MALAT1 knockdown reverses chemoresistance to temozolomide via promoting microRNA-101 in glioblastoma. Cancer Med. 2018, 7, 1404–1415. [Google Scholar] [CrossRef] [Green Version]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef] [Green Version]
- Tiedge, H.; Chen, W.; Brosius, J. Primary structure, neural-specific expression, and dendritic location of human BC200 RNA. J. Neurosci. 1993, 13, 2382–2390. [Google Scholar] [CrossRef] [PubMed]
- Booy, E.P.; McRae, E.K.S.; Howard, R.; Deo, S.R.; Ariyo, E.O.; Dzananovic, E.; Meier, M.; Stetefeld, J.; McKenna, S.A. RNA Helicase Associated with AU-rich Element (RHAU/DHX36) Interacts with the 3’-Tail of the Long Non-coding RNA BC200 (BCYRN1). J. Boil. Chem. 2016, 291, 5355–5372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; Bocker, W.; Brosius, J.; Tiedge, H. Expression of neural BC200 RNA in human tumours. J. Pathol. 1997, 183, 345–351. [Google Scholar] [CrossRef]
- Iacoangeli, A.; Lin, Y.; Morley, E.J.; Muslimov, I.A.; Bianchi, R.; Reilly, J.; Weedon, J.; Diallo, R.; Böcker, W.; Tiedge, H. BC200 RNA in invasive and preinvasive breast cancer. Carcinogenesis 2004, 25, 2125–2133. [Google Scholar] [CrossRef]
- Franken, N.A.; Rodermond, H.M.; Stap, J.; Haveman, J.; van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef]
- Vermes, I.; Haanen, C.; Reutelingsperger, C. Flow cytometry of apoptotic cell death. J. Immunol. Methods 2000, 243, 167–190. [Google Scholar] [CrossRef]
- Arshad, H.; Ahmad, Z.; Hasan, S.H. Gliomas: Correlation of Histologic Grade, Ki67 and p53 Expression with Patient Survival. Asian. Pac. J. Cancer Prev. 2010, 11, 1637–1640. [Google Scholar]
- Mu, N.; Gu, J.; Liu, N.; Xue, X.; Shu, Z.; Zhang, K.; Huang, T.; Chu, C.; Zhang, W.; Gong, L.; et al. PRL-3 is a potential glioblastoma prognostic marker and promotes glioblastoma progression by enhancing MMP7 through the ERK and JNK pathways. Theranostics 2018, 8, 1527–1539. [Google Scholar] [CrossRef]
- Butler, S.J.; Richardson, L.; Farias, N.; Morrison, J.; Coomber, B.L. Characterization of cancer stem cell drug resistance in the human colorectal cancer cell lines HCT116 and SW480. Biochem. Biophys. Res. Commun. 2017, 490, 29–35. [Google Scholar] [CrossRef]
- Yasgar, A.; Titus, S.A.; Wang, Y.; Danchik, C.; Yang, S.-M.; Vasiliou, V.; Jadhav, A.; Maloney, D.J.; Simeonov, A.; Martinez, N.J. A High-Content Assay Enables the Automated Screening and Identification of Small Molecules with Specific ALDH1A1-Inhibitory Activity. PLoS ONE 2017, 12, e0170937. [Google Scholar] [CrossRef]
- Zhou, S.; Schuetz, J.D.; Bunting, K.D.; Colapietro, A.M.; Sampath, J.; Morris, J.J.; Lagutina, I.; Grosveld, G.C.; Osawa, M.; Nakauchi, H.; et al. The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nat. Med. 2001, 7, 1028–1034. [Google Scholar] [CrossRef] [PubMed]
- Schaich, M.; Kestel, L.; Pfirrmann, M.; Robel, K.; Illmer, T.; Kramer, M.; Dill, C.; Ehninger, G.; Schackert, G.; Krex, D. A MDR1 (ABCB1) gene single nucleotide polymorphism predicts outcome of temozolomide treatment in glioblastoma patients. Ann. Oncol. 2009, 20, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Yang, R. Dopamine receptor D1 promotes the proliferation, invasion and migration of gliomas by inhibiting cAMP signaling pathway. Xi bao yu fen zi mian yi xue za zhi = Chin. J. Cell. Mol. Immunolog. 2018, 34, 1116–1121. [Google Scholar]
- Xu, N.; Papagiannakopoulos, T.; Pan, G.; Thomson, J.A.; Kosik, K.S. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 2009, 137, 647–658. [Google Scholar] [CrossRef] [Green Version]
- Kertesz, M.; Iovino, N.; Unnerstall, U.; Gaul, U.; Segal, E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007, 39, 1278–1284. [Google Scholar] [CrossRef]
- Naydenov, E.; Tzekov, C.; Minkin, K.; Nachev, S.; Romansky, K.; Bussarsky, V. Long-term survival with primary glioblastoma multiforme: A clinical study in bulgarian patients. Case Rep. Oncol. 2011, 4, 1–11. [Google Scholar] [CrossRef]
- Bredel, M. Anticancer drug resistance in primary human brain tumors. Brain Res. Rev. 2001, 35, 161–204. [Google Scholar] [CrossRef]
- Tseng, Y.-Y.; Huang, Y.-C.; Yang, T.-C.; Yang, S.-T.; Liu, S.-C.; Chang, T.-M.; Kau, Y.-C.; Liu, S.-J. Concurrent Chemotherapy of Malignant Glioma in Rats by Using Multidrug-Loaded Biodegradable Nanofibrous Membranes. Sci. Rep. 2016, 6, 30630. [Google Scholar] [CrossRef] [Green Version]
- Tivnan, A.; Zakaria, Z.; O’Leary, C.; Kögel, D.; Pokorny, J.L.; Sarkaria, J.N.; Prehn, J.H.; Kögel, N. Inhibition of multidrug resistance protein 1 (MRP1) improves chemotherapy drug response in primary and recurrent glioblastoma multiforme. Front. Mol. Neurosci. 2015, 9, 218. [Google Scholar] [CrossRef] [Green Version]
- Iacoangeli, A.; Adzovic, L.; Chen, E.Q.; Cattie, R.L.; Soff, G.A.; Tiedge, H. Regulatory BC200 RNA in peripheral blood of patients with invasive breast cancer. J. Investig. Med. 2018, 66, 1055–1063. [Google Scholar] [CrossRef]
- Deng, L.; Xiong, P.; Luo, Y.; Bu, X.; Qian, S.; Zhong, W.; Lv, S. Association between IDH1/2 mutations and brain glioma grade. Oncol. Lett. 2018, 16, 5405–5409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stancheva, G.; Goranova, T.; Laleva, M.; Kamenova, M.; Mitkova, A.; Velinov, N.; Poptodorov, G.; Mitev, V.; Kaneva, R.; Gabrovsky, G. IDH1/IDH2 but not TP53 mutations predict prognosis in Bulgarian glioblastoma patients. BioMed Res. Int. 2014, 2014, 6547272014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hata, N.; Hatae, R.; Yoshimoto, K.; Murata, H.; Kuga, D.; Akagi, Y.; Sangatsuda, Y.; Suzuki, S.O.; Iwaki, T.; Mizoguchi, M.; et al. Insular primary glioblastomas with IDH mutations: Clinical and biological specificities. Neuropathology 2017, 37, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, L.; Cui, Y.; Zhang, C.; Chen, H.; Gu, J.; Qian, J.; Luo, C. Increased RLIP76 expression in IDH1 wild-type glioblastoma multiforme is associated with worse prognosis. Oncol. Rep. 2019, 43, 188–200. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Ramena, G.; Elble, R.C. The role of cancer stem cells in relapse of solid tumors. Front. Biosci. (Elite Ed) 2012, 4, 1528–1541. [Google Scholar] [CrossRef] [PubMed]
- Vescovi, A.; Binda, E. Heterogeneity of cancer-Initiating cells within glioblastoma. Front. Biosci. (Schol Ed) 2012, 4, 1235–1248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sang, Y. Lee. Temozolomide resistance in glioblastoma multiforme. Gene Funct. Dis. 2016, 3, 198–210. [Google Scholar]
- Hegi, M.E.; Liu, L.; Herman, J.G.; Stupp, R.; Wick, W.; Weller, M.; Mehta, M.; Gilbert, M.R. Correlation of O6-Methylguanine Methyltransferase (MGMT) Promoter Methylation With Clinical Outcomes in Glioblastoma and Clinical Strategies to Modulate MGMT Activity. J. Clin. Oncol. 2008, 26, 4189–4199. [Google Scholar] [CrossRef] [Green Version]
- Sarkaria, J.N.; Kitange, G.J.; James, C.D.; Plummer, R.; Calvert, H.; Weller, M.; Wick, W. Mechanisms of chemoresistance to alkylating agents in malignant glioma. Clin. Cancer Res. 2008, 14, 2900–2908. [Google Scholar] [CrossRef] [Green Version]
- Yoshimoto, K.; Mizoguchi, M.; Hata, N.; Murata, H.; Hatae, R.; Amano, T.; Nakamizo, A.; Sasaki, T. Complex DNA repair pathways as possible therapeutic targets to overcome temozolomide resistance in glioblastoma. Front. Oncol. 2012, 2, 186. [Google Scholar] [CrossRef] [Green Version]
- Wu, K.; Xu, K.; Liu, K.; Huang, J.; Chen, J.; Zhang, J.; Zhang, N. Long noncoding RNA BC200 regulates cell growth and invasion in colon cancer. Int. J. Biochem. Cell Boil. 2018, 99, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Hou, F.; Feng, J.; Xu, S.; Meng, X. Long non-coding RNA BCYRN1 promotes the proliferation and metastasis of cervical cancer via targeting microRNA-138 in vitro and in vivo. Oncol. Lett. 2018, 15, 5809–5818. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Xu, C.; Ding, B.; Gao, M.; Wei, X.; Ji, N. Long non-coding RNA MALAT1 promotes proliferation and suppresses apoptosis of glioma cells through derepressing Rap1B by sponging miR-101. J. Neuro-Oncol. 2017, 134, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Yan, Y.; Hu, M.; Wang, Y.; Wang, Y.; Dai, Y.; Chen, J.; Di, G.; Chen, X.; Jiang, X. MiR-218 sensitizes glioma cells to apoptosis and inhibits tumorigenicity by regulating ECOP-mediated suppression of NF-κB activity. Neuro-Oncology 2012, 15, 413–422. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Qian, R.; Zhang, J.; Shi, X. MiR-218-5p targets LHFPL3 to regulate proliferation, migration, and epithelial–mesenchymal transitions of human glioma cells. Biosci. Rep. 2019, 39, BSR20180879. [Google Scholar] [CrossRef] [Green Version]
- Riganti, C.; Salaroglio, I.C.; Caldera, V.; Campia, I.; Kopecka, J.; Mellai, M.; Annovazzi, L.; Bosia, A.; Ghigo, D.; Schiffer, D. Temozolomide downregulates P-glycoprotein expression in glioblastoma stem cells by interfering with the Wnt3a/glycogen synthase-3 kinase/β-catenin pathway. Neuro-Oncology 2013, 15, 1502–1517. [Google Scholar] [CrossRef] [Green Version]








| High BC200 n=25 (52%) | Low BC200 n=23 (48%) | p-Value | |
|---|---|---|---|
| Age | |||
| <65 | 15 (60%) | 8 (35%) | 0.0806 |
| ≥65 | 10 (40%) | 15 (65%) | |
| Gender | |||
| male | 14 (56%) | 13 (57%) | 0.971 |
| female | 11 (44%) | 10 (43%) | |
| IDH1 status | |||
| wildtype | 24 (96%) | 17 (74%) | 0.0303 |
| mutation | 1 (4%) | 6 (26%) | |
| P53 status | |||
| wildtype | 9 (36%) | 15 (65%) | 0.0431 |
| mutation | 16 (64%) | 8 (35%) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, Y.-K.; Lin, J.W.; Shih, J.-W.; Chuang, H.-Y.; Fong, I.-H.; Yeh, C.-T.; Lin, C.-M. Targeting BC200/miR218-5p Signaling Axis for Overcoming Temozolomide Resistance and Suppressing Glioma Stemness. Cells 2020, 9, 1859. https://doi.org/10.3390/cells9081859
Su Y-K, Lin JW, Shih J-W, Chuang H-Y, Fong I-H, Yeh C-T, Lin C-M. Targeting BC200/miR218-5p Signaling Axis for Overcoming Temozolomide Resistance and Suppressing Glioma Stemness. Cells. 2020; 9(8):1859. https://doi.org/10.3390/cells9081859
Chicago/Turabian StyleSu, Yu-Kai, Jia Wei Lin, Jing-Wen Shih, Hao-Yu Chuang, Iat-Hang Fong, Chi-Tai Yeh, and Chien-Min Lin. 2020. "Targeting BC200/miR218-5p Signaling Axis for Overcoming Temozolomide Resistance and Suppressing Glioma Stemness" Cells 9, no. 8: 1859. https://doi.org/10.3390/cells9081859
APA StyleSu, Y. -K., Lin, J. W., Shih, J. -W., Chuang, H. -Y., Fong, I. -H., Yeh, C. -T., & Lin, C. -M. (2020). Targeting BC200/miR218-5p Signaling Axis for Overcoming Temozolomide Resistance and Suppressing Glioma Stemness. Cells, 9(8), 1859. https://doi.org/10.3390/cells9081859