PlantMirP2: An Accurate, Fast and Easy-To-Use Program for Plant Pre-miRNA and miRNA Prediction
Abstract
:1. Introduction
2. Materials and Methods
2.1. Datasets and Feature Set
2.2. Performance Evaluation
2.3. SVMs
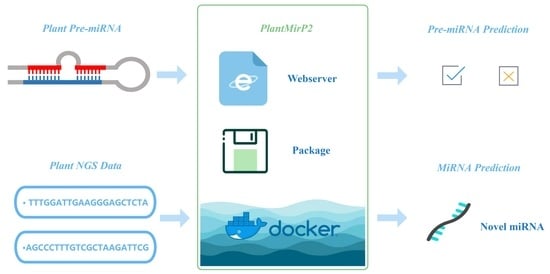
2.4. Implementation of PlantMirP2 Stand-Alone and Web-Server
3. Results
3.1. An Improved Algorithm for the Prediction of Plant Pre-miRNAs
3.2. Prediction for New Plant Pre-miRNAs in miRBase 22.1
3.3. Comparison with miPlantPreMat Based on Dataset of miPlantPreMat
3.4. Comparisons with PlantMiRNAPred, Triplet-SVM and MicroPred Based on Datasets of PlantMiRNAPred
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Teotia, S.; Singh, D.; Tang, G. Technologies to address plant microRNA functions. In Plant microRNAs; Springer: Berlin/Heidelberg, Germany, 2020; pp. 25–43. [Google Scholar]
- Pompili, V.; Piazza, S.; Li, M.; Varotto, C.; Malnoy, M. Transcriptional regulation of MdmiR285N microRNA in apple (Malus x domestica) and the heterologous plant system Arabidopsis thaliana. Hortic. Res. 2020, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Liebsch, D.; Palatnik, J.F. MicroRNA miR396, GRF transcription factors and GIF co-regulators: A conserved plant growth regulatory module with potential for breeding and biotechnology. Curr. Opin. Plant Biol. 2020, 53, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Apostolova, E.; Gozmanova, M.; Nacheva, L.; Ivanova, Z.; Toneva, V.; Minkov, I.; Baev, V.; Yahubyan, G. MicroRNA profiling the resurrection plant Haberlea rhodopensis unveils essential regulators of survival under severe drought. Biol. Plant. 2020, 64, 541–550. [Google Scholar] [CrossRef]
- Niu, Y.; Su, M.; Wu, Y.; Fu, L.; Kang, K.; Li, Q.; Li, L.; Hui, G.; Li, F.; Gou, D. Circulating Plasma miRNAs as Potential Biomarkers of Non–Small Cell Lung Cancer Obtained by High-Throughput Real-Time PCR Profiling. Cancer Epidemiol. Prev. Biomark. 2019, 28, 327–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, S.; Song, X.; Song, W.; Bi, S. An enzyme-free molecular catalytic device: Dynamically self-assembled DNA dendrimers for in situ imaging of microRNAs in live cells. Chem. Sci. 2019, 10, 1651–1658. [Google Scholar] [CrossRef] [Green Version]
- Miller, B.R.; Wei, T.; Fields, C.J.; Sheng, P.; Xie, M. Near-infrared fluorescent northern blot. Rna 2018, 24, 1871–1877. [Google Scholar] [CrossRef]
- Válóczi, A.; Hornyik, C.; Varga, N.; Burgyán, J.; Kauppinen, S.; Havelda, Z. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004, 32, e175. [Google Scholar] [CrossRef] [Green Version]
- Lai, E.C.; Tomancak, P.; Williams, R.W.; Rubin, G.M. Computational identification of Drosophila microRNA genes. Genome Biol. 2003, 4, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Lim, L.P.; Lau, N.C.; Weinstein, E.G.; Abdelhakim, A.; Yekta, S.; Rhoades, M.W.; Burge, C.B.; Bartel, D.P. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003, 17, 991–1008. [Google Scholar] [CrossRef] [Green Version]
- Friedlander, M.R.; Chen, W.; Adamidi, C.; Maaskola, J.; Einspanier, R.; Knespel, S.; Rajewsky, N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008, 26, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Xue, C.; Li, F.; He, T.; Liu, G.-P.; Li, Y.; Zhang, X. Classification of real and pseudo microRNA precursors using local structure-sequence features and support vector machine. BMC Bioinform. 2005, 6, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng, K.L.S.; Mishra, S.K. De novo SVM classification of precursor microRNAs from genomic pseudo hairpins using global and intrinsic folding measures. Bioinformatics 2007, 23, 1321–1330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batuwita, R.; Palade, V. microPred: Effective classification of pre-miRNAs for human miRNA gene prediction. Bioinformatics 2009, 25, 989–995. [Google Scholar] [CrossRef] [Green Version]
- Xuan, P.; Guo, M.; Liu, X.; Huang, Y.; Li, W.; Huang, Y. PlantMiRNAPred: Efficient classification of real and pseudo plant pre-miRNAs. Bioinformatics 2011, 27, 1368–1376. [Google Scholar] [CrossRef] [Green Version]
- Gudyś, A.; Szcześniak, M.W.; Sikora, M.; Makałowska, I. HuntMi: An efficient and taxon-specific approach in pre-miRNA identification. BMC Bioinform. 2013, 14, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Meng, J.; Liu, D.; Sun, C.; Luan, Y. Prediction of plant pre-microRNAs and their microRNAs in genome-scale sequences using structure-sequence features and support vector machine. BMC Bioinform. 2014, 15, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yones, C.; Stegmayer, G.; Milone, D.H. Genome-wide pre-miRNA discovery from few labeled examples. Bioinformatics 2018, 34, 541–549. [Google Scholar] [CrossRef] [Green Version]
- Yao, Y.; Ma, C.; Deng, H.; Liu, Q.; Zhang, J.; Yi, M. plantMirP: An efficient computational program for the prediction of plant pre-miRNA by incorporating knowledge-based energy features. Mol. BioSystems 2016, 12, 3124–3131. [Google Scholar] [CrossRef]
- An, J.; Lai, J.; Lehman, M.L.; Nelson, C.C. miRDeep*: An integrated application tool for miRNA identification from RNA sequencing data. Nucleic Acids Res. 2013, 41, 727–737. [Google Scholar] [CrossRef]
- Yang, X.; Li, L. miRDeep-P: A computational tool for analyzing the microRNA transcriptome in plants. Bioinformatics 2011, 27, 2614–2615. [Google Scholar] [CrossRef]
- Kuang, Z.; Wang, Y.; Li, L.; Yang, X. miRDeep-P2: Accurate and fast analysis of the microRNA transcriptome in plants. Bioinformatics 2019, 35, 2521–2522. [Google Scholar] [CrossRef]
- Mathelier, A.; Carbone, A. MIReNA: Finding microRNAs with high accuracy and no learning at genome scale and from deep sequencing data. Bioinformatics 2010, 26, 2226–2234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, J.; Lai, J.; Sajjanhar, A.; Lehman, M.L.; Nelson, C.C. miRPlant: An integrated tool for identification of plant miRNA from RNA sequencing data. BMC Bioinform. 2014, 15, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Lei, J.; Sun, Y. miR-PREFeR: An accurate, fast and easy-to-use plant miRNA prediction tool using small RNA-Seq data. Bioinformatics 2014, 30, 2837–2839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evers, M.; Huttner, M.; Dueck, A.; Meister, G.; Engelmann, J.C. miRA: Adaptable novel miRNA identification in plants using small RNA sequencing data. BMC Bioinform. 2015, 16, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Wang, H.; Yao, Y.; Yi, M. PlantMirP-Rice: An Efficient Program for Rice Pre-miRNA Prediction. Genes 2020, 11, 662. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Zhang, H.; Deng, H. milRNApredictor: Genome-free prediction of fungi milRNAs by incorporating k-mer scheme and distance-dependent pair potential. Genomics 2020, 112, 2233–2240. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2012, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed] [Green Version]










| Positive Dataset | Species | PlantMirP (Release 21) | PlantMirP2 (Release 22.1) |
|---|---|---|---|
| Training | Arabidopsis thaliana | 325 | 326 |
| Glycine max | 573 | 684 | |
| Oryza sativa | 592 | 604 | |
| Physcomitrella patens | 229 | 247 | |
| Medicago truncatula | 672 | 672 | |
| Sorghum bicolor | 205 | 205 | |
| Arabidopsis lyrata | 205 | 205 | |
| Zea mays | 166 | 168 | |
| Solanum lycopersicum | 77 | 112 | |
| Testing | Remaining plant species | 3865 | 5323 |
| NO. | Features | Description | Origin |
|---|---|---|---|
| 1–34 | Knowledge-based energy score1 | Calculated using the position-specific contact potentials of 2-mer pairs. | riceMirP |
| 35–39 | Knowledge-based energy score2 | Calculated using the distance-specific contact potentials of k-mer pairs (k = 1~5). | plantMirP |
| 40–49 | The ratio of unpaired nucleotide in sub-region 1–10 | The secondary structure was divided into 10 parts and the ratio of unpaired nucleotide in each part was calculated. | plantMirP |
| 50 | the size of biggest bulge | The size of biggest bulge in secondary structure. A bugle contains at least three adjacent unpaired nucleotides. | plantMirP |
| 51 | n_stems/L | n_stems denotes the number of stems. A stem contains at least three continuous base pairs. L is the length of sequence. | plantMirP |
| 52 | n_loops/L | n_loops denotes the number of loops. | plantMirP |
| 53 | %(|G| + |C|) | (|G| + |C|)/L × 100. Here |X| denotes the number of base X in sequence. | miPred |
| 54–69 | %XY | |XY|/(L − 1) × 100. |XY| is number of dinucleotide XY in sequence. | miPred |
| 70 | dG = MFE/L | MFE is minimum of free energy of the secondary structure. | miPred |
| 71 | MFE1 | (MFE/L)/%(|G| + |C|) | miPred |
| 72 | MFE2 | (MFE/L)/n_stems | miPred |
| 73 | dP = tot_bases/L | tot_bases is number of base pairs in the secondary structure. | miPred |
| 74 | MFE3 | (MFE/L)/n_loops | microPred |
| 75–77 | |X − Y|/L | |X − Y| is the number of base pairs, (X − Y)∈[(A − U), (G − C), (G − U)] | microPred |
| 78–80 | %(X − Y)/n_stems | %(X − Y) = |X − Y|/n_stems × 100 | microPred |
| 81 | Avg_bp_stem1 | tot_bases/n_stems | microPred |
| 82 | pb/nb | paired nucleotide/unpaired nucleotide | miRD |
| 83 | MCPN | Maximum of consecutive paired nucleotides. | ZmirP |
| 84 | n_bugles/L | n_bulges is the total number of bulges in the secondary structure. | ZmirP |
| 85 | Avg_bp_stem2 | The ratio of number of base pairs to n_stems | ZmirP |
| 86 | MFE4 | dG/tot_bases | ZmirP |
| 87 | MFE5 | dG/n_bugles | ZmirP |
| 88–167 | k-mer features | k-mer features (k = 2 & 3). | milRP |
| 168–193 | Knowledge-based energy score3 | Calculated using the distance-dependent k-mer pair potential (k = 1–3 and Nbins = 20). | milRP |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, D.; Yao, Y.; Yi, M. PlantMirP2: An Accurate, Fast and Easy-To-Use Program for Plant Pre-miRNA and miRNA Prediction. Genes 2021, 12, 1280. https://doi.org/10.3390/genes12081280
Fan D, Yao Y, Yi M. PlantMirP2: An Accurate, Fast and Easy-To-Use Program for Plant Pre-miRNA and miRNA Prediction. Genes. 2021; 12(8):1280. https://doi.org/10.3390/genes12081280
Chicago/Turabian StyleFan, Dashuai, Yuangen Yao, and Ming Yi. 2021. "PlantMirP2: An Accurate, Fast and Easy-To-Use Program for Plant Pre-miRNA and miRNA Prediction" Genes 12, no. 8: 1280. https://doi.org/10.3390/genes12081280
APA StyleFan, D., Yao, Y., & Yi, M. (2021). PlantMirP2: An Accurate, Fast and Easy-To-Use Program for Plant Pre-miRNA and miRNA Prediction. Genes, 12(8), 1280. https://doi.org/10.3390/genes12081280