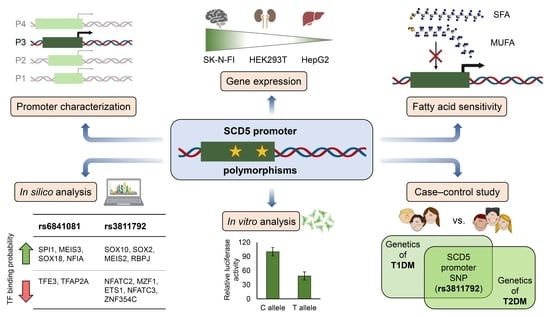
A Single Nucleotide Polymorphism (rs3811792) Affecting Human SCD5 Promoter Activity Is Associated with Diabetes Mellitus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Web-Based Tools for in Silico Transcription Factor Binding Analysis
2.3. Reporter Plasmid Construction and Mutagenesis
2.4. Cell Culture and Transfection
2.5. Cell Treatment
2.6. Preparation of Cell Lysates
2.7. Luciferase Assay
2.8. qPCR Analysis
2.9. Subjects
2.10. Genotyping
2.11. Statistical Analysis
3. Results
3.1. Cell Type-Specific Promoter Activity of Human Stearoyl-CoA Desaturase-5
3.2. Fatty Acid Insensitive Promoter Activity of Stearoyl-CoA Desaturase-5
3.3. Effect of rs3811792 Polymorphism on Stearoyl-CoA Desaturase-5 Promoter Activity
3.4. Effect of rs6841081 and rs3811792 Promoter Polymorphisms on Transcription Factor Binding Sites in Silico
3.5. Association of rs3811792 Polymorphism with T1DM and T2DM
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Saeedi, P.; Petersohn, I.; Salpea, P.; Malanda, B.; Karuranga, S.; Unwin, N.; Colagiuri, S.; Guariguata, L.; Motala, A.A.; Ogurtsova, K.; et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Res. Clin. Pract. 2019, 157, 107843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, H.; Saeedi, P.; Karuranga, S.; Pinkepank, M.; Ogurtsova, K.; Duncan, B.B.; Stein, C.; Basit, A.; Chan, J.C.N.; Mbanya, J.C.; et al. IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract. 2022, 183, 109119. [Google Scholar] [CrossRef]
- DiMeglio, L.A.; Evans-Molina, C.; Oram, R.A. Type 1 diabetes. Lancet 2018, 391, 2449–2462. [Google Scholar] [CrossRef]
- Wu, Y.; Ding, Y.; Tanaka, Y.; Zhang, W. Risk factors contributing to type 2 diabetes and recent advances in the treatment and prevention. Int. J. Med. Sci. 2014, 11, 1185–1200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robertson, C.C.; Rich, S.S. Genetics of type 1 diabetes. Curr. Opin. Genet. Dev. 2018, 50, 7–16. [Google Scholar] [CrossRef]
- Laakso, M. Biomarkers for type 2 diabetes. Mol. Metab. 2019, 27, S139–S146. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.C.; Philipson, L.H. Update on diabetes classification. Med. Clin. North Am. 2015, 99, 1–16. [Google Scholar] [CrossRef]
- Wilkin, T.J. The accelerator hypothesis: A review of the evidence for insulin resistance as the basis for type I as well as type II diabetes. Int. J. Obes. 2009, 33, 716–726. [Google Scholar] [CrossRef] [Green Version]
- Ravaut, G.; Légiot, A.; Bergeron, K.F.; Mounier, C. Monounsaturated Fatty Acids in Obesity-Related Inflammation. Int. J. Mol. Sci. 2020, 22, 330. [Google Scholar] [CrossRef]
- Enoch, H.G.; Catalá, A.; Strittmatter, P. Mechanism of rat liver microsomal stearyl-CoA desaturase. Studies of the substrate specificity, enzyme-substrate interactions, and the function of lipid. J. Biol. Chem. 1976, 251, 5095–5103. [Google Scholar] [CrossRef]
- Popeijus, H.E.; Saris, W.H.; Mensink, R.P. Role of stearoyl-CoA desaturases in obesity and the metabolic syndrome. Int. J. Obes. 2008, 32, 1076–1082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ntambi, J.M.; Miyazaki, M. Recent insights into stearoyl-CoA desaturase-1. Curr. Opin. Lipidol. 2003, 14, 255–261. [Google Scholar] [CrossRef] [PubMed]
- AM, A.L.; Syed, D.N.; Ntambi, J.M. Insights into Stearoyl-CoA Desaturase-1 Regulation of Systemic Metabolism. Trends Endocrinol. Metab. TEM 2017, 28, 831–842. [Google Scholar] [CrossRef]
- Jeyakumar, S.M.; Vajreswari, A. Stearoyl-CoA desaturase 1: A potential target for non-alcoholic fatty liver disease?-perspective on emerging experimental evidence. World J. Hepatol. 2022, 14, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Tabaczar, S.; Wołosiewicz, M.; Filip, A.; Olichwier, A.; Dobrzyń, P. The role of stearoyl-CoA desaturase in the regulation of cardiac metabolism. Postep. Biochem. 2018, 64, 183–189. [Google Scholar] [CrossRef]
- Mauvoisin, D.; Mounier, C. Hormonal and nutritional regulation of SCD1 gene expression. Biochimie 2011, 93, 78–86. [Google Scholar] [CrossRef]
- Sampath, H.; Ntambi, J.M. Polyunsaturated fatty acid regulation of genes of lipid metabolism. Annu. Rev. Nutr. 2005, 25, 317–340. [Google Scholar] [CrossRef]
- Peter, A.; Weigert, C.; Staiger, H.; Machicao, F.; Schick, F.; Machann, J.; Stefan, N.; Thamer, C.; Häring, H.U.; Schleicher, E. Individual stearoyl-coa desaturase 1 expression modulates endoplasmic reticulum stress and inflammation in human myotubes and is associated with skeletal muscle lipid storage and insulin sensitivity in vivo. Diabetes 2009, 58, 1757–1765. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.N.; Pariza, M.W.; Ntambi, J.M. Conjugated linoleic acid decreases hepatic stearoyl-CoA desaturase mRNA expression. Biochem. Biophys. Res. Commun. 1998, 248, 817–821. [Google Scholar] [CrossRef]
- Waters, K.M.; Miller, C.W.; Ntambi, J.M. Localization of a polyunsaturated fatty acid response region in stearoyl-CoA desaturase gene 1. Biochim. Et Biophys. Acta 1997, 1349, 33–42. [Google Scholar] [CrossRef]
- Ide, T.; Shimano, H.; Yoshikawa, T.; Yahagi, N.; Amemiya-Kudo, M.; Matsuzaka, T.; Nakakuki, M.; Yatoh, S.; Iizuka, Y.; Tomita, S.; et al. Cross-talk between peroxisome proliferator-activated receptor (PPAR) alpha and liver X receptor (LXR) in nutritional regulation of fatty acid metabolism. II. LXRs suppress lipid degradation gene promoters through inhibition of PPAR signaling. Mol. Endocrinol. 2003, 17, 1255–1267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, H.; Sakaki, K.; Mihara, K. Ubiquitin-proteasome-dependent degradation of mammalian ER stearoyl-CoA desaturase. J. Cell Sci. 2006, 119, 2342–2353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tibori, K.; Orosz, G.; Zámbó, V.; Szelényi, P.; Sarnyai, F.; Tamási, V.; Rónai, Z.; Mátyási, J.; Tóth, B.; Csala, M.; et al. Molecular Mechanisms Underlying the Elevated Expression of a Potentially Type 2 Diabetes Mellitus Associated SCD1 Variant. Int. J. Mol. Sci. 2022, 23, 6221. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ge, L.; Tran, T.; Stenn, K.; Prouty, S.M. Isolation and characterization of the human stearoyl-CoA desaturase gene promoter: Requirement of a conserved CCAAT cis-element. Biochem. J. 2001, 357, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ge, L.; Parimoo, S.; Stenn, K.; Prouty, S.M. Human stearoyl-CoA desaturase: Alternative transcripts generated from a single gene by usage of tandem polyadenylation sites. Biochem. J. 1999, 340 Pt 1, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Beiraghi, S.; Zhou, M.; Talmadge, C.B.; Went-Sumegi, N.; Davis, J.R.; Huang, D.; Saal, H.; Seemayer, T.A.; Sumegi, J. Identification and characterization of a novel gene disrupted by a pericentric inversion inv(4)(p13.1q21.1) in a family with cleft lip. Gene 2003, 309, 11–21. [Google Scholar] [CrossRef]
- Wang, J.; Yu, L.; Schmidt, R.E.; Su, C.; Huang, X.; Gould, K.; Cao, G. Characterization of HSCD5, a novel human stearoyl-CoA desaturase unique to primates. Biochem. Biophys. Res. Commun. 2005, 332, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zou, X.; Chang, Q.; Zhang, Y.; Li, Y.; Zhang, L.; Huang, J.; Liang, B. The evolutionary pattern and the regulation of stearoyl-CoA desaturase genes. BioMed Res. Int. 2013, 2013, 856521. [Google Scholar] [CrossRef] [Green Version]
- Minville-Walz, M.; Gresti, J.; Pichon, L.; Bellenger, S.; Bellenger, J.; Narce, M.; Rialland, M. Distinct regulation of stearoyl-CoA desaturase 1 gene expression by cis and trans C18:1 fatty acids in human aortic smooth muscle cells. Genes Nutr. 2012, 7, 209–216. [Google Scholar] [CrossRef] [Green Version]
- Jacobs, A.A.; van Baal, J.; Smits, M.A.; Taweel, H.Z.; Hendriks, W.H.; van Vuuren, A.M.; Dijkstra, J. Effects of feeding rapeseed oil, soybean oil, or linseed oil on stearoyl-CoA desaturase expression in the mammary gland of dairy cows. J. Dairy Sci. 2011, 94, 874–887. [Google Scholar] [CrossRef]
- Antal, O.; Péter, M.; Hackler, L., Jr.; Mán, I.; Szebeni, G.; Ayaydin, F.; Hideghéty, K.; Vigh, L.; Kitajka, K.; Balogh, G.; et al. Lipidomic analysis reveals a radiosensitizing role of gamma-linolenic acid in glioma cells. Biochim. Biophys. Acta 2015, 1851, 1271–1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burhans, M.S.; Flowers, M.T.; Harrington, K.R.; Bond, L.M.; Guo, C.A.; Anderson, R.M.; Ntambi, J.M. Hepatic oleate regulates adipose tissue lipogenesis and fatty acid oxidation. J. Lipid Res. 2015, 56, 304–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Igal, R.A.; Sinner, D.I. Stearoyl-CoA desaturase 5 (SCD5), a Delta-9 fatty acyl desaturase in search of a function. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2021, 1866, 158840. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Luo, N.; Liu, L.; Cui, H.; Li, J.; Xiang, H.; Kang, H.; Li, H.; Zhao, G. Identification of the molecular regulation of differences in lipid deposition in dedifferentiated preadipocytes from different chicken tissues. BMC Genom. 2021, 22, 232. [Google Scholar] [CrossRef] [PubMed]
- Sinner, D.I.; Kim, G.J.; Henderson, G.C.; Igal, R.A. StearoylCoA desaturase-5: A novel regulator of neuronal cell proliferation and differentiation. PLoS ONE 2012, 7, e39787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Sun, S.; Zhang, Y.; Wang, X.; Li, Q. Identification of Scd5 as a functional regulator of visceral fat deposition and distribution. iScience 2022, 25, 103916. [Google Scholar] [CrossRef] [PubMed]
- Castro-Mondragon, J.A.; Riudavets-Puig, R.; Rauluseviciute, I.; Lemma, R.B.; Turchi, L.; Blanc-Mathieu, R.; Lucas, J.; Boddie, P.; Khan, A.; Manosalva Pérez, N.; et al. JASPAR 2022: The 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022, 50, D165–D173. [Google Scholar] [CrossRef]
- Farré, D.; Roset, R.; Huerta, M.; Adsuara, J.E.; Roselló, L.; Albà, M.M.; Messeguer, X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003, 31, 3651–3653. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.; Huang, C.H. LASAGNA-Search 2.0: Integrated transcription factor binding site search and visualization in a browser. Bioinformatics 2014, 30, 1923–1925. [Google Scholar] [CrossRef] [Green Version]
- Moore, C.M.; Jacobson, S.A.; Fingerlin, T.E. Power and Sample Size Calculations for Genetic Association Studies in the Presence of Genetic Model Misspecification. Hum. Hered. 2019, 84, 256–271. [Google Scholar] [CrossRef]
- Carninci, P.; Sandelin, A.; Lenhard, B.; Katayama, S.; Shimokawa, K.; Ponjavic, J.; Semple, C.A.; Taylor, M.S.; Engström, P.G.; Frith, M.C.; et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006, 38, 626–635. [Google Scholar] [CrossRef] [PubMed]
- Roongta, U.V.; Pabalan, J.G.; Wang, X.; Ryseck, R.P.; Fargnoli, J.; Henley, B.J.; Yang, W.P.; Zhu, J.; Madireddi, M.T.; Lawrence, R.M.; et al. Cancer cell dependence on unsaturated fatty acids implicates stearoyl-CoA desaturase as a target for cancer therapy. Mol. Cancer Res. MCR 2011, 9, 1551–1561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murakami, A.; Nagao, K.; Juni, N.; Hara, Y.; Umeda, M. An N-terminal di-proline motif is essential for fatty acid-dependent degradation of Δ9-desaturase in Drosophila. J. Biol. Chem. 2017, 292, 19976–19986. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Yin, X.; Mai, H.; Li, G.; Lin, Z.; Jie, W.; Li, K.; Zhou, H.; Wei, S.; Hu, L.; et al. SCD rs41290540 single-nucleotide polymorphism modifies miR-498 binding and is associated with a decreased risk of coronary artery disease. Mol. Genet. Genom. Med. 2020, 8, e1136. [Google Scholar] [CrossRef] [Green Version]
- Guerrero-Ros, I.; Clement, C.C.; Reynolds, C.A.; Patel, B.; Santambrogio, L.; Cuervo, A.M.; Macian, F. The negative effect of lipid challenge on autophagy inhibits T cell responses. Autophagy 2020, 16, 223–238. [Google Scholar] [CrossRef]
- Lengi, A.J.; Corl, B.A. Comparison of pig, sheep and chicken SCD5 homologs: Evidence for an early gene duplication event. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2008, 150, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Green, C.D.; Olson, L.K. Modulation of palmitate-induced endoplasmic reticulum stress and apoptosis in pancreatic beta-cells by stearoyl-CoA desaturase and Elovl6. Am. J. Physiol. Endocrinol. Metab. 2011, 300, E640–E649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwai, T.; Kume, S.; Chin-Kanasaki, M.; Kuwagata, S.; Araki, H.; Takeda, N.; Sugaya, T.; Uzu, T.; Maegawa, H.; Araki, S.I. Stearoyl-CoA Desaturase-1 Protects Cells against Lipotoxicity-Mediated Apoptosis in Proximal Tubular Cells. Int. J. Mol. Sci. 2016, 17, 1868. [Google Scholar] [CrossRef] [Green Version]
- Sarnyai, F.; Donko, M.B.; Matyasi, J.; Gor-Nagy, Z.; Marczi, I.; Simon-Szabo, L.; Zambo, V.; Somogyi, A.; Csizmadia, T.; Low, P.; et al. Cellular toxicity of dietary trans fatty acids and its correlation with ceramide and diglyceride accumulation. Food Chem. Toxicol. 2019, 124, 324–335. [Google Scholar] [CrossRef] [Green Version]
- Sarnyai, F.; Somogyi, A.; Gor-Nagy, Z.; Zambo, V.; Szelenyi, P.; Matyasi, J.; Simon-Szabo, L.; Kereszturi, E.; Toth, B.; Csala, M. Effect of cis- and trans-Monounsaturated Fatty Acids on Palmitate Toxicity and on Palmitate-induced Accumulation of Ceramides and Diglycerides. Int. J. Mol. Sci. 2020, 21, 2626. [Google Scholar] [CrossRef]
- Zambo, V.; Simon-Szabo, L.; Sarnyai, F.; Matyasi, J.; Gor-Nagy, Z.; Somogyi, A.; Szelenyi, P.; Kereszturi, E.; Toth, B.; Csala, M. Investigation of the putative rate-limiting role of electron transfer in fatty acid desaturation using transfected HEK293T cells. FEBS Lett. 2020, 594, 530–539. [Google Scholar] [CrossRef] [PubMed]
- Zambo, V.; Simon-Szabo, L.; Szelenyi, P.; Kereszturi, E.; Banhegyi, G.; Csala, M. Lipotoxicity in the liver. World J. Hepatol. 2013, 5, 550–557. [Google Scholar] [CrossRef] [PubMed]





| SNP ID | Position | Allele | All Hits (above 1% Relative Score) | At Least 15% Relative Score Difference between Alleles | Relative Score Grater than 80% at Least for One Allele |
|---|---|---|---|---|---|
| rs6841081 | −254 | G | 56,353 | 438 | 6 |
| T | 56,356 | ||||
| rs3811792 | −316 | C | 56,359 | 372 | 9 |
| T | 56,362 |
| Name | TF ID | Strand | Relative Score (%) | ||
|---|---|---|---|---|---|
| G Allele | T Allele | Difference | |||
| SPI1 | MA0080.1 | - | 61.12 | 81.85 | 20.73 |
| MEIS3 | MA0775.1 | - | 64.05 | 81.49 | 17.44 |
| SOX18 | MA1563.1 | + | 67.49 | 83.78 | 16.28 |
| NFIA | MA0670.1 | + | 66.42 | 82.29 | 15.87 |
| TFE3 | MA0831.1 | - | 80.86 | 64.07 | −16.79 |
| TFAP2A | MA0003.1 | - | 86.43 | 67.53 | −18.90 |
| Name | TF ID | Strand | Relative Score (%) | ||
|---|---|---|---|---|---|
| C Allele | T Allele | Difference | |||
| SOX10 | MA0442.1 | + | 61.69 | 80.51 | 18.83 |
| SOX2 | MA0143.4 | - | 69.17 | 85.12 | 15.95 |
| MEIS2 | MA0774.1 | + | 68.63 | 83.88 | 15.25 |
| RBPJ | MA1116.1 | - | 72.83 | 87.87 | 15.04 |
| NFATC2 | MA0152.1 | + | 91.46 | 75.36 | −16.10 |
| ZNF354C | MA0130.1 | + | 86.87 | 68.98 | −17.89 |
| NFATC3 | MA0625.2 | - | 93.30 | 74.36 | −18.94 |
| MZF1 | MA0056.1 | - | 81.31 | 61.97 | −19.34 |
| ETS1 | MA0098.1 | + | 93.09 | 71.50 | −21.60 |
| Control (N = 350) | T1DM (N = 145) | T2DM (N = 253) | |||||
| rs6841081 | N | % | N | % | N | % | |
| GG | 342 | 98 | 141 | 97 | 243 | 96 | |
| GT | 8 | 2 | 4 | 3 | 10 | 4 | |
| TT | 0 | 0 | 0 | 0 | 0 | 0 | |
| χ2 | p = 0.7555 | p = 0.1119 | |||||
| MAF (T) | 8 | 1 | 4 | 1 | 10 | 2 | |
| OR with 95% CI | 1.2128 (0.3594–4.0922) | 1.7593 (0.6844–4.5224) | |||||
| Control (N = 350) | T1DM (N = 143) | T2DM (N = 248) | |||||
| rs3811792 | N | % | N | % | N | % | |
| CC | 248 | 71 | 97 | 68 | 162 | 65 | |
| CT | 94 | 27 | 33 | 23 | 79 | 32 | |
| TT | 8 | 2 | 13 | 9 | 7 | 3 | |
| χ2 | p = 0.0029 | p = 0.0114 | |||||
| MAF (T) | 110 | 16 | 59 | 21 | 93 | 19 | |
| OR with 95% CI | 4.275 (1.7318–10.5527) | 1.2907 (0.9108–1.8291) | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zámbó, V.; Orosz, G.; Szabó, L.; Tibori, K.; Sipeki, S.; Molnár, K.; Csala, M.; Kereszturi, É. A Single Nucleotide Polymorphism (rs3811792) Affecting Human SCD5 Promoter Activity Is Associated with Diabetes Mellitus. Genes 2022, 13, 1784. https://doi.org/10.3390/genes13101784
Zámbó V, Orosz G, Szabó L, Tibori K, Sipeki S, Molnár K, Csala M, Kereszturi É. A Single Nucleotide Polymorphism (rs3811792) Affecting Human SCD5 Promoter Activity Is Associated with Diabetes Mellitus. Genes. 2022; 13(10):1784. https://doi.org/10.3390/genes13101784
Chicago/Turabian StyleZámbó, Veronika, Gabriella Orosz, Luca Szabó, Kinga Tibori, Szabolcs Sipeki, Krisztina Molnár, Miklós Csala, and Éva Kereszturi. 2022. "A Single Nucleotide Polymorphism (rs3811792) Affecting Human SCD5 Promoter Activity Is Associated with Diabetes Mellitus" Genes 13, no. 10: 1784. https://doi.org/10.3390/genes13101784
APA StyleZámbó, V., Orosz, G., Szabó, L., Tibori, K., Sipeki, S., Molnár, K., Csala, M., & Kereszturi, É. (2022). A Single Nucleotide Polymorphism (rs3811792) Affecting Human SCD5 Promoter Activity Is Associated with Diabetes Mellitus. Genes, 13(10), 1784. https://doi.org/10.3390/genes13101784