A Rearrangement of the Mitochondrial Genes of Centipedes (Arthropoda, Myriapoda) with a Phylogenetic Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Taxon Sampling and Mitochondrial DNA Sequencing
2.2. Gene Annotation and Secondary Structure Prediction
2.3. Sequence Alignment and Phylogenetic Analyses
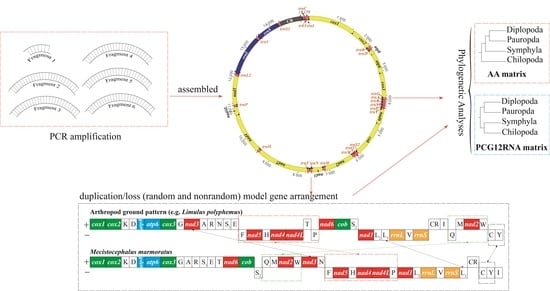
3. Results and Discussion
3.1. Organization of the Mitogenome
3.2. Transfer RNAs
3.3. Phylogenetic Analyses
3.4. Evolution of Gene Rearrangements in the Mitochondrial Genome of Centipedes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Minelli, A. The Myriapoda, Volume 1, Chapter: Chilopoda. In The Myriapoda; Minelli, A., Ed.; Brill: Leiden, The Netherlands, 2011; pp. 1–20. [Google Scholar]
- Edgecombe, G.D.; Giribet, G. Evolutionary biology of centipedes (Myriapoda: Chilopoda). Annu. Rev. Entomol. 2007, 52, 151–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- So, W.L.; Nong, W.Y.; Xie, Y.C.; Baril, T.; Ma, H.Y.; Qu, Z.; Haimovitz, J.; Swale, T.; Gaitan-Espitia, J.D.; Lau, K.F.; et al. Myriapod genomes reveal ancestral horizontal gene transfer and hormonal gene loss in millipedes. Nat. Commun. 2022, 13, 3010. [Google Scholar] [CrossRef]
- Arthur, W.; Chipman, A.D. The centipede Strigamia maritima: What it can tell us about the development and evolution of segmentation. Bioessays 2005, 27, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Brena, C.; Akam, M. The embryonic development of the centipede Strigamia maritima. Dev. Biol. 2012, 363, 290–307. [Google Scholar] [CrossRef] [Green Version]
- Green, J.; Akam, M. Evolution of the pair rule gene network: Insights from a centipede. Dev. Biol. 2013, 382, 235–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ax, P. Ein Lehrbuch der Phylogenetischen Systematik; Gustav Fischer Verlag: Stuttgart, Germany, 1999; p. 384. [Google Scholar]
- Bäcker, H.; Fanenbruck, M.; Wägele, J.W. A forgotten homology supporting the monophyly of Tracheata: The subcoxa of insects and myriapods revisited. Zool. Anz. J. Comp. Zool. 2008, 247, 185–207. [Google Scholar] [CrossRef]
- Bitsch, C.; Bitsch, J. Phylogenetic relationships of basal hexapods among the mandibulate arthropods: A cladistic analysis based on comparative morphological characters. Zool. Scr. 2004, 33, 511–550. [Google Scholar] [CrossRef]
- Boudreaux, H. Significance of intersegmental tendon system in arthropod phylogeny and a monophyletic classification of Arthropoda. In Arthropod Phylogeny; Van Nostrand Reinhold: New York, NY, USA, 1979; pp. 551–586. [Google Scholar]
- Chipman, A.D.; Ferrier, D.E.; Brena, C.; Qu, J.; Hughes, D.S.; Schröder, R.; Torres-Oliva, M.; Znassi, N.; Jiang, H.; Almeida, F.C.; et al. The first myriapod genome sequence reveals conservative arthropod gene content and genome organisation in the centipede Strigamia maritima. PLoS Biol. 2014, 12, e1002005. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.; Sun, H.; Guo, H.; Pan, D.; Qian, C.; Hao, S.; Zhou, K. The complete mitochondrial genome of Pauropus longiramus (Myriapoda: Pauropoda): Implications on early diversification of the myriapods revealed from comparative analysis. Gene 2012, 505, 57–65. [Google Scholar] [CrossRef]
- Dong, Y.; Zhu, L.; Bai, Y.; Ou, Y.; Wang, C. Complete mitochondrial genomes of two flat-backed millipedes by next-generation sequencing (Diplopoda, Polydesmida). ZooKeys 2016, 637, 1–20. [Google Scholar] [CrossRef]
- Gai, Y.H.; Song, D.X.; Sun, H.Y.; Zhou, K.Y. Myriapod monophyly and relationships among myriapod classes based on nearly complete 28S and 18S rDNA sequences. Zool. Sci. 2006, 23, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, B.G.; Jamieson, J.B.G. The Ultrastructure and Phylogeny of Insect Spermatozoa; Cambridge University Press: New York, NY, USA, 1987. [Google Scholar]
- Regier, J.C.; Shultz, J.W.; Ganley, A.R.; Hussey, A.; Shi, D.; Ball, B.; Zwick, A.; Stajich, J.E.; Cummings, M.P.; Martin, J.W.; et al. Resolving arthropod phylogeny: Exploring phylogenetic signal within 41 kb of protein-coding nuclear gene sequence. Syst. Biol. 2008, 57, 920–938. [Google Scholar] [CrossRef] [PubMed]
- Regier, J.C.; Shultz, J.W.; Zwick, A.; Hussey, A.; Ball, B.; Wetzer, R.; Martin, J.W.; Cunningham, C.W. Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature 2010, 463, 1079–1083. [Google Scholar] [CrossRef] [PubMed]
- Regier, J.C.; Wilson, H.M.; Shultz, J.W. Phylogenetic analysis of Myriapoda using three nuclear protein-coding genes. Mol. Phylogenet. Evol. 2005, 34, 147–158. [Google Scholar] [CrossRef]
- Szucsich, N.U.; Bartel, D.; Blanke, A.; Böhm, A.; Donath, A.; Fukui, M.; Grove, S.; Liu, S.; Macek, O.; Machida, R.; et al. Four myriapod relatives—But who are sisters? No end to debates on relationships among the four major myriapod subgroups. BMC Evol. Biol. 2020, 20, 144. [Google Scholar] [CrossRef]
- Zuo, Q.; Zhang, Z.; Shen, Y. Novel mitochondrial gene rearrangements pattern in the millipede Polydesmus sp. GZCS-2019 and phylogenetic analysis of the Myriapoda. Ecol. Evol. 2022, 12, e8764. [Google Scholar] [CrossRef]
- Benavides, L.R.; Edgecombe, G.D.; Giribet, G. Re-evaluating and dating myriapod diversification with phylotranscriptomics under a regime of dense taxon sampling. Mol. Phylogenet. Evol. 2022, 178, 107621. [Google Scholar] [CrossRef]
- Wang, J.J.; Bai, Y.; Zhao, H.; Mu, R.; Dong, Y. Reinvestigating the phylogeny of Myriapoda with more extensive taxon sampling and novel genetic perspective. PeerJ 2021, 9, e12691. [Google Scholar] [CrossRef]
- Edgecombe, G.D.; Wilson, G.D.; Colgan, D.J.; Gray, M.R.; Cassis, G. Arthropod cladistics: Combined analysis of histone H3 and U2 snRNA sequences and morphology. Cladistics 2000, 16, 155–203. [Google Scholar] [CrossRef]
- Fernández, R.; Edgecombe, G.D.; Giribet, G. Phylogenomics illuminates the backbone of the Myriapoda Tree of Life and reconciles morphological and molecular phylogenies. Sci. Rep. 2018, 8, 83. [Google Scholar] [CrossRef]
- Pocock, R.I. Contributions to our knowledge of the arthropod fauna of the West Indies. Part III. Diplopoda and Malacopoda, with a supplement on the Arachnida of the class Pedipalpi. Zool. J. Linn. Soc. 1894, 24, 473–544. [Google Scholar] [CrossRef]
- Tiegs, O.W. The development and affinities of the Pauropoda, based on a study of Pauropus silvaticus. J. Cell Sci. 1947, 3, 275–336. [Google Scholar] [CrossRef]
- Fernández, R.; Edgecombe, G.D.; Giribet, G. Exploring phylogenetic relationships within Myriapoda and the effects of matrix composition and occupancy on phylogenomic reconstruction. Syst. Biol. 2016, 65, 871–889. [Google Scholar] [CrossRef]
- Edgecombe, G.D. Morphological data, extant Myriapoda, and the myriapod stem-group. Contrib. Zool. 2004, 73, 207–252. [Google Scholar] [CrossRef] [Green Version]
- Edgecombe, G.D.; Giribet, G.; Wheeler, W.C. Phylogeny of Henicopidae (Chilopoda: Lithobiomorpha): A combined analysis of morphology and five molecular loci. Syst. Entomol. 2002, 27, 31–64. [Google Scholar] [CrossRef] [Green Version]
- Giribet, G.; Carranza, S.; Riutort, M.; Baguna, J.; Ribera, C. Internal phylogeny of the Chilopoda (Myriapoda, Arthropoda) using complete 18S rDNA and partial 28S rDNA sequences. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1999, 354, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Benavides, L.R.; Jiang, C.; Giribet, G. Mimopidae is the sister group to all other scolopendromorph centipedes (Chilopoda, Scolopendromorpha): A phylotranscriptomic approach. Org. Divers. Evol. 2021, 21, 591–598. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [Green Version]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [Green Version]
- Song, F.; Li, H.; Shao, R.; Shi, A.; Bai, X.; Zheng, X.; Heiss, E.; Cai, W. Rearrangement of mitochondrial tRNA genes in flat bugs (Hemiptera: Aradidae). Sci. Rep. 2016, 6, 25725. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, X.; Garzón-Orduña, I.J.; Winterton, S.L.; Yan, Y.; Aspöck, U.; Aspöck, H.; Yang, D. Mitochondrial phylogenomics illuminates the evolutionary history of Neuropterida. Cladistics 2017, 33, 617–636. [Google Scholar] [CrossRef] [PubMed]
- Dowton, M.; Cameron, S.L.; Dowavic, J.I.; Austin, A.D.; Whiting, M.F. Characterization of 67 mitochondrial tRNA gene rearrangements in the Hymenoptera suggests that mitochondrial tRNA gene position is selectively neutral. Mol. Biol. Evol. 2009, 26, 1607–1617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz-Mena, A.; Mora, P.; Montiel, E.E.; Palomeque, T.; Lorite, P. Complete Nucleotide Sequence of the Mitogenome of Tapinoma ibericum (Hymenoptera: Formicidae: Dolichoderinae), Gene Organization and Phylogenetics Implications for the Dolichoderinae Subfamily. Genes 2022, 13, 1325. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Xu, J.J.; Hao, S.J.; Sun, H.Y. The complete mitochondrial genome of the giant pill millipede, Sphaerotheriidae sp. (Myriapoda: Diplopoda: Sphaerotheriida). Mitochondrial DNA 2012, 23, 333–335. [Google Scholar] [CrossRef] [PubMed]
- Gai, Y.; Ma, H.; Ma, J.; Li, C.; Yang, Q. The complete mitochondrial genome of Scolopocryptops sp. (Chilopoda: Scolopendromorpha: Scolopocryptopidae). Mitochondrial DNA 2014, 25, 192–193. [Google Scholar] [CrossRef]
- Lavrov, D.V.; Brown, W.M.; Boore, J.L. A novel type of RNA editing occurs in the mitochondrial tRNAs of the centipede Lithobius forficatus. Proc. Natl. Acad. Sci. USA 2000, 97, 13738–13742. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Fang, Y.; Cao, G.; Shen, C.; Liu, H.; Ruan, H. The Complete Mitochondrial Genome of Spirobolus bungii (Diplopoda, Spirobolidae): The First Sequence for the Genus Spirobolus. Genes 2022, 13, 1587. [Google Scholar] [CrossRef]
- Hu, C.; Wang, S.; Huang, B.; Liu, H.; Xu, L.; Hu, Z.; Liu, Y. The complete mitochondrial genome sequence of Scolopendra mutilans L. Koch, 1878 (Scolopendromorpha, Scolopendridae), with a comparative analysis of other centipede genomes. ZooKeys 2020, 925, 73–88. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Qi, Y.; Tian, X. Analysis of mitochondrial genome of Scolopendra subspinipes dehaani. Tianjin J. Tradit. Chin. Med. 2018, 35, 225–229. [Google Scholar]
- Robertson, H.E.; Lapraz, F.; Rhodes, A.C.; Telford, M.J. The complete mitochondrial genome of the geophilomorph centipede Strigamia maritima. PLoS ONE 2015, 10, e0121369. [Google Scholar] [CrossRef] [PubMed]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Gai, Y.; Song, D.; Sun, H.; Yang, Q.; Zhou, K. The complete mitochondrial genome of Symphylella sp. (Myriapoda: Symphyla): Extensive gene order rearrangement and evidence in favor of Progoneata. Mol. Phylogenet. Evol. 2008, 49, 574–585. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef] [PubMed]
- Laslett, D.; Canbäck, B. ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 2008, 24, 172–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Ranwez, V.; Douzery, E.J.; Cambon, C.; Chantret, N.; Delsuc, F. MACSE v2: Toolkit for the alignment of coding sequences accounting for frameshifts and stop codons. Mol. Biol. Evol. 2018, 35, 2582–2584. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef] [Green Version]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [Green Version]
- Huelsenbeck, J.P.; Ronquist, F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Nabhan, A.R.; Sarkar, I.N. The impact of taxon sampling on phylogenetic inference: A review of two decades of controversy. Brief. Bioinform. 2012, 13, 122–134. [Google Scholar] [CrossRef] [Green Version]
- Negrisolo, E.; Minelli, A.; Valle, G. The mitochondrial genome of the house centipede Scutigera and the monophyly versus paraphyly of myriapods. Mol. Biol. Evol. 2004, 21, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Lavrov, D.V.; Boore, J.L.; Brown, W.M. Complete mtDNA sequences of two millipedes suggest a new model for mitochondrial gene rearrangements: Duplication and nonrandom loss. Mol. Biol. Evol. 2002, 19, 163–169. [Google Scholar] [CrossRef] [Green Version]
- Park, S.J.; Choi, E.H.; Hwang, J.S.; Hwang, U.W. The complete mitochondrial genome of a centipede Bothropolys sp. (Chilopoda, Lithobiomorpha, Lithobiidae). Mitochondrial DNA Part A 2016, 27, 2268–2269. [Google Scholar]
- Gai, Y.; Ma, H.; Sun, X.; Ma, J.; Li, C.; Yang, Q. The complete mitochondrial genome of Cermatobius longicornis (Chilopoda: Lithobiomorpha: Henicopidae). Mitochondrial DNA 2013, 24, 331–332. [Google Scholar] [CrossRef]
- Woo, H.J.; Lee, Y.S.; Park, S.J.; Lim, J.T.; Jang, K.H.; Choi, E.H.; Choi, Y.G.; Hwang, U.W. Complete mitochondrial genome of a troglobite millipede Antrokoreana gracilipes (Diplopoda, Juliformia, Julida), and juliformian phylogeny. Mol. Cells 2007, 23, 182–191. [Google Scholar]
- Brewer, M.S.; Swafford, L.; Spruill, C.L.; Bond, J.E. Arthropod phylogenetics in light of three novel millipede (Myriapoda: Diplopoda) mitochondrial genomes with comments on the appropriateness of mitochondrial genome sequence data for inferring deep level relationships. PLoS ONE 2013, 8, e68005. [Google Scholar] [CrossRef]
- Podsiadlowski, L.; Kohlhagen, H.; Koch, M. The complete mitochondrial genome of Scutigerella causeyae (Myriapoda: Symphyla) and the phylogenetic position of Symphyla. Mol. Phylogenet. Evol. 2007, 45, 251–260. [Google Scholar] [CrossRef]
- Park, S.J.; Lee, Y.S.; Hwang, U.W. The complete mitochondrial genome of the sea spider Achelia bituberculata (Pycnogonida, Ammotheidae): Arthropod ground pattern of gene arrangement. BMC Genom. 2007, 8, 343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masta, S.E.; Boore, J.L. Parallel evolution of truncated transfer RNA genes in arachnid mitochondrial genomes. Mol. Biol. Evol. 2008, 25, 949–959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavrov, D.V.; Boore, J.L.; Brown, W.M. The complete mitochondrial DNA sequence of the horseshoe crab Limulus polyphemus. Mol. Biol. Evol. 2000, 17, 813–824. [Google Scholar] [CrossRef] [PubMed]
- Shingate, P.; Ravi, V.; Prasad, A.; Tay, B.H.; Venkatesh, B. Chromosome-level genome assembly of the coastal horseshoe crab (Tachypleus gigas). Mol. Ecol. Resour. 2020, 20, 1748–1760. [Google Scholar] [CrossRef]
- Wilson, K.; Cahill, V.; Ballment, E.; Benzie, J. The complete sequence of the mitochondrial genome of the crustacean Penaeus monodon: Are malacostracan crustaceans more closely related to insects than to branchiopods? Mol. Biol. Evol. 2000, 17, 863–874. [Google Scholar] [CrossRef] [Green Version]
- Lavrov, D.V.; Brown, W.M.; Boore, J.L. Phylogenetic position of the Pentastomida and (pan) crustacean relationships. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2004, 271, 537–544. [Google Scholar] [CrossRef]
- Podsiadlowski, L. The mitochondrial genome of the bristletail Petrobius brevistylis (Archaeognatha: Machilidae). Insect Mol. Biol. 2006, 15, 253–258. [Google Scholar] [CrossRef]
- Cook, C.E.; Yue, Q.; Akam, M. Mitochondrial genomes suggest that hexapods and crustaceans are mutually paraphyletic. Proc. R. Soc. B Biol. Sci. 2005, 272, 1295–1304. [Google Scholar] [CrossRef] [Green Version]
- Clary, D.O.; Wolstenholme, D.R. The ribosomal RNA genes of Drosophila mitochondrial DNA. Nucleic Acids Res. 1985, 13, 4029–4045. [Google Scholar] [CrossRef] [Green Version]
- Carapelli, A.; Nardi, F.; Dallai, R.; Boore, J.; Lio, P.; Frati, F. Relationships between hexapods and crustaceans based on four mitochondrial genes. Crustacean Issues 2005, 16, 295. [Google Scholar]
- Webster, B.L.; Copley, R.R.; Jenner, R.A.; Mackenzie-Dodds, J.A.; Bourlat, S.J.; Rota-Stabelli, O.; Littlewood, D.; Telford, M.J. Mitogenomics and phylogenomics reveal priapulid worms as extant models of the ancestral Ecdysozoan. Evol. Dev. 2006, 8, 502–510. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Lan, H.; Xu, W.; Chen, Y.N.; Wu, H.; Jiang, H.M.; Wang, J.C.; Wu, Y.B.; Liu, H. Two complete mitochondrial genomes in Scolopendra and a comparative analysis of tRNA rearrangements in centipedes. Mol. Biol. Rep. 2022, 49, 6173–6180. [Google Scholar] [CrossRef] [PubMed]





| Primer Name | Nucleotide Sequence (5′-3′) | PCR Amplification Product Length | Reference |
|---|---|---|---|
| CO1CF | GCACGTCTACAAATCATAAAGATATTGG | 0.7 kb | [46] |
| CO1CR | TAAACTTCAGGGTGACCGAAAAATCA | ||
| Lco1 | TTATAATTTTTTTTATAGTGATACC | 3.7 kb | [12] |
| CO3R | ACATCTACAAAATGTCAGTATCA | [47] | |
| Dco3F | TATCATCCTATCAATGATGACGAGA | 3.7 kb | [12] |
| Dn4R | ATTTATGATTACCTAAGGCTCATG | ||
| Dn4F | ATGAACAACAGAAGAATAAGC | 2.9 kb | |
| Hcob | GCAAATAAAAAATATCATTCTGGTTG | ||
| DcobF | ATAATAACCGCCTTCTTGGGAT | 3.4 kb | |
| D12SR | CTGTTTCTGAATCGATATTCCACGTTT | ||
| D12SF | ATAATAGGGTATCTAATCCTAGTCT | 2.7 kb | |
| Dco1R | ATGGGGGATATACGGTCCATCCGG |
| Species | Mitochondrial Genome | PCGs | ||||
|---|---|---|---|---|---|---|
| A + T | AT Skew | GC Skew | A + T | AT Skew | GC Skew | |
| Bothropolys sp. | 70.6 | 0.07 | −0.31 | 68.6 | −0.13 | −0.06 |
| C.s longicornis | 63.4 | 0.09 | −0.32 | 60.5 | −0.17 | −0.05 |
| Lithobius forficatus | 67.9 | 0.09 | −0.27 | 65.7 | −0.14 | −0.03 |
| M. marmoratus | 69.5 | 0.12 | −0.33 | 67.1 | −0.11 | −0.02 |
| S. maritima | 64.1 | 0.22 | −0.33 | 62.1 | −0.12 | −0.02 |
| S. dehaani | 74.1 | 0.13 | −0.34 | 59.6 | −0.12 | −0.01 |
| S. mutilans | 78.8 | 0.05 | −0.26 | 77.2 | −0.12 | −0.04 |
| S. subspinipes | 72.7 | 0.06 | −0.33 | 71.6 | −0.14 | 0 |
| Scolopendra morsitans | 72.8 | 0.02 | −0.38 | 71.6 | −0.14 | −0.02 |
| Scolopocryptops sp. | 71.6 | 0.03 | −0.31 | 70.4 | −0.14 | −0.01 |
| S. coleoptrata | 69.4 | 0.04 | −0.31 | 68.3 | −0.14 | −0.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.-J.; Bai, Y.; Dong, Y. A Rearrangement of the Mitochondrial Genes of Centipedes (Arthropoda, Myriapoda) with a Phylogenetic Analysis. Genes 2022, 13, 1787. https://doi.org/10.3390/genes13101787
Wang J-J, Bai Y, Dong Y. A Rearrangement of the Mitochondrial Genes of Centipedes (Arthropoda, Myriapoda) with a Phylogenetic Analysis. Genes. 2022; 13(10):1787. https://doi.org/10.3390/genes13101787
Chicago/Turabian StyleWang, Jia-Jia, Yu Bai, and Yan Dong. 2022. "A Rearrangement of the Mitochondrial Genes of Centipedes (Arthropoda, Myriapoda) with a Phylogenetic Analysis" Genes 13, no. 10: 1787. https://doi.org/10.3390/genes13101787
APA StyleWang, J. -J., Bai, Y., & Dong, Y. (2022). A Rearrangement of the Mitochondrial Genes of Centipedes (Arthropoda, Myriapoda) with a Phylogenetic Analysis. Genes, 13(10), 1787. https://doi.org/10.3390/genes13101787