Gene Network Analysis for Osteoporosis, Sarcopenia, Diabetes, and Obesity in Human Mesenchymal Stromal Cells
Abstract
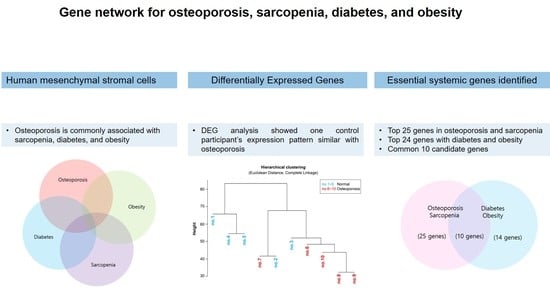
:1. Introduction
2. Materials and Methods
2.1. Study Subjects
2.2. Human MSC Isolation and Culture
2.3. RNA Isolation
2.4. RNA Library Preparation and Sequencing
2.5. Analysis of Gene Expression
2.5.1. Hierarchical Clustering
2.5.2. Network Analysis
2.6. Statistical Analysis
3. Results
3.1. Clinical Characteristics of Study Participants
3.2. Comparison of Gene Expression Patterns between Control and Osteoporosis Groups
3.3. RNA Analysis of Bone Remodeling Epigenome
3.4. Genetic Relationship among Osteoporosis, Obesity and Diabetes
3.5. Common Related Genes in Osteoporosis, Sarcopenia, Diabetes, and Obesity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sambrook, P.; Cooper, C. Osteoporosis. Lancet 2006, 367, 2010–2018. [Google Scholar] [CrossRef]
- Ensrud, K.E.; Crandall, C.J. Osteoporosis. Ann. Intern. Med. 2017, 167, ITC17–ITC32. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.; Hassan, Z. Osteoporosis: Current Concepts. Joints 2018, 6, 122–127. [Google Scholar]
- Lin, J.T.; Lane, J.M. Osteoporosis: A Review. Clin. Orthop. Relat. Res. 2004, 425, 126–134. [Google Scholar] [CrossRef]
- Obesity and Overweight: Factsheet. Available online: http://www.who.int/mediacentre/factsheets/fs311/en/ (accessed on 22 November 2018).
- Hatim, B.; Amanda, C.M.; Bradley, A.; Ou, C.N.; Mercedes, M.G.; Lourdes, G.S.; Fernando, C.; Francisco, J.T.; Inmaculada, M.S.; Manuel, M.G. Transcriptional Analysis of FOXO1, C/EBP-α and PPAR-γ2 Genes and Their Association with Obesity-Related Insulin Resistance. Genes 2019, 10, 706. [Google Scholar]
- Hegele, R.A.; Cao, H.; Frankowski, C.; Mathews, S.T.; Leff, T. PPARG F388L, a Transactivation-Deficient Mutant, in Familial Partial Lipodystrophy. Diabetes 2002, 51, 3586–3590. [Google Scholar] [CrossRef] [Green Version]
- Wallenius, V.; Wallenius, K.; Ahrén, B.; Rudling, M.; Carlsten, H.; Dickson, S.L.; Ohlsson, C.; Jansson, J.O. Interleukin-6-Deficient Mice Develop Mature-Onset Obesity. Nat. Med. 2002, 8, 75–79. [Google Scholar] [CrossRef]
- Lee, N.K.; Sowa, H.; Hinoi, E.; Ferron, M.; Ahn, J.D.; Confavreux, C.; Dacquin, R.; Mee, P.J.; McKee, M.D.; Jung, D.Y.; et al. Endocrine Regulation of Energy Metabolism by the Skeleton. Cell 2007, 130, 456–469. [Google Scholar] [CrossRef] [Green Version]
- Salem, H.K.; Thiemermann, C. Mesenchymal Stromal Cells: Current Understanding and Clinical Status. Stem Cells 2010, 28, 585–596. [Google Scholar] [CrossRef] [Green Version]
- Chamberlain, G.; Fox, J.; Ashton, B.; Middleton, J. Concise Review: Mesenchymal Stem Cells: Their Phenotype, Differentiation Capacity, Immunological Features, and Potential for Homing. Stem Cells 2007, 25, 2739–2749. [Google Scholar] [CrossRef] [Green Version]
- Benisch, P.; Schilling, T.; Klein-Hitpass, L.; Frey, S.P.; Seefried, L.; Raaijmakers, N.; Krug, M.; Regensburger, M.; Zeck, S.; Schinke, T.; et al. The Transcriptional Profile of Mesenchymal Stem Cell Populations in Primary Osteoporosis Is Distinct and Shows Overexpression of Osteogenic Inhibitors. PLoS ONE 2012, 7, e45142. [Google Scholar] [CrossRef] [PubMed]
- James, A.W. Review of Signaling Pathways Governing MSC Osteogenic and Adipogenic Differentiation. Scientifica 2013, 2013, 684736. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Song, I.; Jin, Y.; Jin, H.S.; Ji, H.M.; Jeong, S.Y.; Won, Y.Y.; Chung, Y.S. Transcriptional Profiling of Human Femoral Mesenchymal Stem Cells in Osteoporosis and Its Association with Adipogenesis. Gene 2017, 632, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, A.V. Diabetes Mellitus: Does It Affect Bone? Calcif. Tissue Int. 2003, 73, 515–519. [Google Scholar] [CrossRef]
- Leidig-Bruckner, G.; Ziegler, R. Diabetes Mellitus a Risk for Osteoporosis? Exp. Clin. Endocrinol. Diabetes 2001, 109 (Suppl. 2), S493–S514. [Google Scholar] [CrossRef]
- Inzerillo, A.M.; Epstein, S. Osteoporosis and Diabetes Mellitus. Rev. Endocr. Metab. Disord. 2004, 5, 261–268. [Google Scholar] [CrossRef]
- Thrailkill, K.M.; Lumpkin, C.K., Jr.; Bunn, R.C.; Kemp, S.F.; Fowlkes, J.L. Is Insulin an Anabolic Agent in Bone? Dissecting the Diabetic Bone for Clues. Am. J. Physiol. Endocrinol. Metab. 2005, 289, E735–E745. [Google Scholar] [CrossRef] [Green Version]
- Mani, A.; Radhakrishnan, J.; Wang, H.; Mani, A.; Mani, M.A.; Nelson-Williams, C.; Carew, K.S.; Mane, S.; Najmabadi, H.; Wu, D.; et al. LRP6 Mutation in a Family with Early Coronary Disease and Metabolic Risk Factors. Science 2007, 315, 1278–1282. [Google Scholar] [CrossRef] [Green Version]
- Oh, S.W.; Shin, S.A.; Yun, Y.H.; Yoo, T.; Huh, B.Y. Cut-Off Point of BMI and Obesity-Related Comorbidities and Mortality in Middle-Aged Koreans. Obes. Res. 2004, 12, 2031–2040. [Google Scholar] [CrossRef] [Green Version]
- Angelopoulou, M.; Novelli, E.; Grove, J.E.; Rinder, H.M.; Civin, C.; Cheng, L.; Krause, D.S. Cotransplantation of Human Mesenchymal Stem Cells Enhances Human Myelopoiesis and Megakaryocytopoiesis in NOD/SCID Mice. Exp. Hematol. 2003, 31, 413–420. [Google Scholar] [CrossRef]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering Splice Junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript Assembly and Quantification by RNA-Seq Reveals Unannotated Transcripts and Isoform Switching During Cell Differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal Criteria for Defining Multipotent Mesenchymal Stromal Cells. The International Society for Cellular Therapy Position Statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Golpanian, S.; Wolf, A.; Hatzistergos, K.E.; Hare, J.M. Rebuilding the Damaged Heart: Mesenchymal Stem Cells, Cell-Based Therapy, and Engineered Heart Tissue. Physiol. Rev. 2016, 96, 1127–1168. [Google Scholar] [CrossRef] [PubMed]
- Ozsolak, F.; Milos, P.M. RNA Sequencing: Advances, Challenges and Opportunities. Nat. Rev. Genet. 2011, 12, 87–98. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A Revolutionary Tool for Transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Marioni, J.C.; Mason, C.E.; Mane, S.M.; Stephens, M.; Gilad, Y. RNA-Seq: An Assessment of Technical Reproducibility and Comparison with Gene Expression Arrays. Genome Res. 2008, 18, 1509–1517. [Google Scholar] [CrossRef] [Green Version]
- Martin, J.A.; Wang, Z. Next-Generation Transcriptome Assembly. Nat. Rev. Genet. 2011, 12, 671–682. [Google Scholar] [CrossRef]
- Seeliger, C.; Karpinski, K.; Haug, A.T.; Vester, H.; Schmitt, A.; Bauer, J.S.; van Griensven, M. Five Freely Circulating miRNAs and Bone Tissue miRNAs Are Associated with Osteoporotic Fractures. J. Bone Miner. Res. 2014, 29, 1718–1728. [Google Scholar] [CrossRef]
- Lee, K.G.; Lee, G.B.; Yang, J.S.; Moon, M.H. Perturbations of Lipids and Oxidized Phospholipids in Lipoproteins of Patients with Postmenopausal Osteoporosis Evaluated by Asymmetrical Flow Field-Flow Fractionation and Nanoflow UHPLC–ESI–MS/MS. Antioxidants 2020, 9, 46. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Yang, M.M.; Li, Y.B.; Liu, G.D.; Teng, Y.; Liu, X.M. Association of CFH and CFB Gene Polymorphisms with Retinopathy in type 2 Diabetic Patients. Mediat. Inflamm. 2013, 2013, 748435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsunaga, H.; Iwashita, M.; Shinjo, T.; Yamashita, A.; Tsuruta, M.; Nagasaka, S.; Taniguchi, A.; Fukushima, M.; Watanabe, N.; Nishimura, F. Adipose Tissue Complement Factor B Promotes Adipocyte Maturation. Biochem. Biophys. Res. Commun. 2018, 495, 740–748. [Google Scholar] [CrossRef]
- Ortega, F.J.; Moreno-Navarrete, J.M.; Sabater, M.; Ricart, W.; Frühbeck, G.; Fernández-Real, J.M. Circulating Glucagon Is Associated with Inflammatory Mediators in Metabolically Compromised Subjects. Eur. J. Endocrinol. 2011, 165, 639–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grey, A.; Mitnick, M.A.; Masiukiewicz, U.; Sun, B.H.; Rudikoff, S.; Jilka, R.L.; Manolagas, S.C.; Insogna, K. A Role for Interleukin-6 in Parathyroid Hormone-Induced Bone Resorption In Vivo. Endocrinology 1999, 140, 4683–4690. [Google Scholar] [CrossRef] [PubMed]
- Ha, J.; Choi, H.S.; Lee, Y.; Kwon, H.J.; Song, Y.W.; Kim, H.H. CXC Chemokine Ligand 2 Induced by Receptor Activator of NF-κB Ligand Enhances Osteoclastogenesis. J. Immunol. 2010, 184, 4717–4724. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, X.; Li, Y.; Chen, A.; Liang, W.; Liang, G.; Huang, B.; Li, Q.; Jin, D. CXCL2 Attenuates Osteoblast Differentiation by Inhibiting the ERK1/2 Signaling Pathway. J. Cell Sci. 2019, 132, jcs230490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rouault, C.; Pellegrinelli, V.; Schilch, R.; Cotillard, A.; Poitou, C.; Tordjman, J.; Sell, H.; Clément, K.; Lacasa, D. Roles of Chemokine ligand-2 (CXCL2) and Neutrophils in Influencing Endothelial Cell Function and Inflammation of Human Adipose Tissue. Endocrinology 2013, 154, 1069–1079. [Google Scholar] [CrossRef] [Green Version]
- Hwang, J.Y.; Lee, S.H.; Kim, G.S.; Koh, J.M.; Go, M.J.; Kim, Y.J.; Kim, H.C.; Kim, T.H.; Hong, J.M.; Park, E.K.; et al. HSD11B1 Polymorphisms Predicted Bone Mineral Density and Fracture Risk in Postmenopausal Women Without a Clinically Apparent Hypercortisolemia. Bone 2009, 45, 1098–1103. [Google Scholar] [CrossRef]
- Gathercole, L.L.; Stewart, P.M. Targeting the Pre-Receptor Metabolism of Cortisol as a Novel Therapy in Obesity and Diabetes. J. Steroid Biochem. Mol. Biol. 2010, 122, 21–27. [Google Scholar] [CrossRef]
- Nair, S.; Lee, Y.H.; Lindsay, R.S.; Walker, B.R.; Tataranni, P.A.; Bogardus, C.; Baier, L.J.; Permana, P.A. 11β-Hydroxysteroid Dehydrogenase Type 1: Genetic Polymorphisms Are Associated with Type 2 Diabetes in Pima Indians Independently of Obesity and Expression in Adipocyte and Muscle. Diabetologia 2004, 47, 1088–1095. [Google Scholar] [CrossRef] [Green Version]
- Ascenzi, F.; Barberi, L.; Dobrowolny, G.; Villa Nova Bacurau, A.; Nicoletti, C.; Rizzuto, E.; Rosenthal, N.; Scicchitano, B.M.; Musarò, A. Effects of IGF-1 isoforms on muscle growth and sarcopenia. Aging Cell 2019, 18, e12954. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, Y.F.; Wen, Y.X.; Tan, Y.; Qin, J.; Jiang, H.Q.; Magdalou, J.; Chne, L.B.; Wang, H. Intrauterine Programming of Glucocorticoid–Insulin-Like Growth Factor-1 Axis–Mediated Developmental Origin of Osteoporosis Susceptibility in Female Offspring Rats with Prenatal Caffeine Exposure. Am. J. Pathol. 2018, 188, 2863–2876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Su, J.; Wang, M.M. Changes of serum IGF-1 and ET-1 levels in patients with osteoporosis and its clinical significance. Pak. J. Med. Sci. 2019, 35, 691–695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moyer-Mileur, L.J.; Slater, H.; Jordan, K.C.; Murray, M.A. IGF-1 and IGF-Binding Proteins and Bone Mass, Geometry, and Strength: Relation to Metabolic Control in Adolescent Girls with type 1 Diabetes. J. Bone Miner. Res. 2008, 23, 1884–1891. [Google Scholar] [CrossRef] [PubMed]
- Ardawi, M.S.; Akhbar, D.H.; AlShaikh, A.; Ahmed, M.M.; Qari, M.H.; Rouzi, A.A.; Ali, A.Y.; Abdulrafee, A.A.; Saeda, M.Y. Increased Serum Sclerostin and Decreased Serum IGF-1 Are Associated with Vertebral Fractures Among Postmenopausal Women with type-2 Diabetes. Bone 2013, 56, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Mediero, A.; Ramkhelawon, B.; Perez-Aso, M.; Moore, K.J.; Cronstein, B.N. Netrin-1 Is a Critical Autocrine/Paracrine Factor for Osteoclast Differentiation. J. Bone Miner. Res. 2015, 30, 837–854. [Google Scholar] [CrossRef] [Green Version]
- Sato, T.; Kokabu, S.; Enoki, Y.; Hayashi, N.; Matsumoto, M.; Nakahira, M.; Sugasawa, M.; Yoda, T. Functional Roles of netrin-1 in Osteoblast Differentiation. In Vivo 2017, 31, 321–328. [Google Scholar] [CrossRef] [Green Version]
- Frank, G.R.; Fox, J.; Candela, N.; Jovanovic, Z.; Bochukova, E.; Levine, J.; Papenhausen, P.R.; O’Rahilly, S.; Farooqi, I.S. Severe Obesity and Diabetes Insipidus in a Patient with PCSK1 Deficiency. Mol. Genet. Metab. 2013, 110, 191–194. [Google Scholar] [CrossRef]
- Ridderstråle, M.; Johansson, L.E.; Rastam, L.; Lindblad, U. Increased Risk of Obesity Associated with the Variant Allele of the PPARGC1A Gly482Ser Polymorphism in Physically Inactive Elderly Men. Diabetologia 2006, 49, 496–500. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Feng, Z.; Wang, X.; Zeng, M.; Liu, J.; Han, S.; Xu, J.; Chen, L.; Cao, K.; Long, J.; et al. SIRT3/SOD2 Maintains Osteoblast Differentiation and Bone Formation by Regulating Mitochondrial Stress. Cell Death Differ. 2018, 25, 229–240. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Lee, Y.D.; Kim, H.J.; Lee, Z.H.; Kim, H.H. SOD2 and Sirt3 Control Osteoclastogenesis by Regulating Mitochondrial ROS. J. Bone Miner. Res. 2017, 32, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.M.; Russell, J.W.; Sullivan, K.A.; Backus, C.; Hayes, J.M.; McLean, L.L.; Feldman, E.L. SOD2 Protects Neurons from Injury in Cell Culture and Animal Models of Diabetic Neuropathy. Exp. Neurol. 2007, 208, 216–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madkour, M.I.; El-Serafi, A.T.; Jahrami, H.A.; Sherif, N.M.; Hassan, R.E.; Awadallah, S.; Faris, M.A.E. Ramadan Diurnal Intermittent Fasting Modulates SOD2, TFAM, Nrf2, and Sirtuins (SIRT1, SIRT3) Gene Expressions in Subjects with Overweight and Obesity. Diabetes Res. Clin. Pract. 2019, 155, 107801. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Jian, J.; Abramson, S.B.; Huang, X. Inhibitory Effects of Iron on Bone Morphogenetic Protein 2–Induced Osteoblastogenesis. J. Bone Miner. Res. 2011, 26, 1188–1196. [Google Scholar] [CrossRef]
- Ellervik, C.; Mandrup-Poulsen, T.; Andersen, H.U.; Tybjærg-Hansen, A.; Frandsen, M.; Birgens, H.; Nordestgaard, B.G. Elevated Transferrin Saturation and Risk of Diabetes: Three Population-Based Studies. Diabetes Care 2011, 34, 2256–2258. [Google Scholar] [CrossRef] [Green Version]
- Rosen, C.J.; Klibanski, A. Bone, Fat, and Body Composition: Evolving Concepts in the Pathogenesis of Osteoporosis. Am. J. Med. 2009, 122, 409–414. [Google Scholar] [CrossRef]
- Al Anouti, F.; Taha, Z.; Shamim, S.; Khalaf, K.; Al Kaabi, L.; Alsafar, H. An Insight into the Paradigms of Osteoporosis: From Genetics to Biomechanics. Bone Rep. 2019, 11, 100216. [Google Scholar] [CrossRef]
- Li, B.; Wang, Y.; Liu, Y.; Ma, J.; Li, Y. Altered Gene Expression Involved in Insulin Signaling Pathway in type II Diabetic Osteoporosis Rats Model. Endocrine 2013, 43, 136–146. [Google Scholar] [CrossRef]




| (a) | ||||||||||||
| Subjects | Bone Mineral Density | |||||||||||
| no. | Age (yrs) | Height (cm) | Weight (kg) | Lumbar Spine (g/cm2) | Femur Neck (g/cm2) | Total Hip (g/cm2) | Whole BMD (g/cm2) | ASMI (kg/m2) | LBM (kg) | BMI (kg/m2) | Diabetes | |
| Control (n = 5) | 1 | 70.0 | 154.5 | 75.2 | 1.023 | 0.865 | 0.920 | 1.068 | 6.728 | 37.088 | 32 | No |
| 2 | 42.0 | 164.0 | 88.1 | 1.279 | 1.023 | 1.102 | 1.277 | 7.745 | 47.568 | 33 | No | |
| 3 | 64.0 | 150.1 | 55.2 | 1.016 | 1.022 | 0.989 | 0.970 | 6.133 | 31.741 | 25 | No | |
| 4 | 55.0 | 172.0 | 70.6 | 1.269 | 1.102 | 1.157 | 1.380 | 6.946 | 47.762 | 24 | No | |
| 5 | 71.0 | 150.0 | 70.3 | 1.016 | 0.905 | 1.134 | 1.201 | 6.938 | 39.237 | 31 | No | |
| Mean | 60.4 | 158.12 | 71.9 | 1.121 | 0.983 | 1.060 | 1.179 | 6.898 | 40.679 | 28.77 | ||
| Osteoporosis (n = 5) | 6 | 85.0 | 160.0 | 49.1 | 0.713 | 0.519 | 0.625 | 0.760 | 4.339 | 30.890 | 19 | Yes |
| 7 | 71.0 | 151.0 | 46.6 | 0.775 | 0.461 | 0.520 | 0.743 | 5.472 | 33.145 | 20 | Yes | |
| 8 | 79.0 | 152.0 | 59.0 | 0.922 | 0.558 | 0.579 | - | - | - | 26 | Yes | |
| 9 | 80.0 | 151.0 | 54.0 | 0.810 | 0.636 | 0.623 | 0.878 | 4.613 | 28.831 | 24 | Yes | |
| 10 | 72.0 | 141.0 | 54.8 | 0.794 | 0.696 | 0.728 | 0.836 | 5.384 | 28.872 | 28 | No | |
| Mean | 77.4 | 151.0 | 52.7 | 0.803 | 0.574 | 0.615 | 0.804 | 4.952 | 30.435 | 23.28 | ||
| (b) | ||||||||||||
| Subjects | Bone Mineral Density | |||||||||||
| no. | Lumbar Spine (g/cm2) | Femur Neck (g/cm2) | Total Hip (g/cm2) | Whole BMD (g/cm2) | ||||||||
| Control (n = 5) | 1 | 1.024 | 0.866 | 0.921 | 1.073 | |||||||
| 2 | 1.139 | 0.909 | 0.978 | 1.131 | ||||||||
| 3 | 1.082 | 1.076 | 1.048 | 1.045 | ||||||||
| 4 | 1.010 | 0.892 | 0.928 | 1.107 | ||||||||
| 5 | 1.084 | 0.960 | 1.194 | 1.278 | ||||||||
| Mean | 1.068 | 0.940 | 1.014 | 1.127 | ||||||||
| Osteoporosis (n = 5) | 6 | 0.632 | 0.453 | 0.554 | 0.678 | |||||||
| 7 | 0.828 | 0.504 | 0.567 | 0.804 | ||||||||
| 8 | 0.960 | 0.589 | 0.613 | |||||||||
| 9 | 0.863 | 0.679 | 0.670 | 1.098 | ||||||||
| 10 | 0.995 | 0.860 | 0.906 | 0.841 | ||||||||
| Mean | 0.856 | 0.617 | 0.662 | 0.855 | ||||||||
| p-value | 0.0141 | 0.0039 | 0.0026 | 0.0194 | ||||||||
| Gene Symbol | no. 1 | no. 2 | no. 3 | no. 4 | no. 5 |
|---|---|---|---|---|---|
| ACKR3 | 6.855 | 1.439 | 1.303 | 3.895 | 15.063 |
| AGT | −2.107 | 1.005 | 4.004 | −1.015 | 3.397 |
| BCL2 | −1.472 | 1.542 | 2.091 | 1.187 | −1.068 |
| BDKRB2 | 27.36 | 3.53 | 1.683 | 12.459 | 17.274 |
| BMP2 | 8.682 | −1.198 | 1.278 | 2.626 | 6.76 |
| C3AR1 | 1.315 | 3.404 | 1.158 | 2.263 | 1.045 |
| C5AR1 | −2.64 | 1.496 | 1.305 | −1.334 | −2.939 |
| CCL5 | 5.637 | 2.635 | 1.199 | 1.268 | 4.472 |
| CFB | 9.544 | 1.4 | 1.361 | 2.173 | 3.967 |
| CSF1 | 4.442 | 2.414 | 1.685 | 2.024 | 3.503 |
| CXCL2 | 9.746 | 1.278 | 2.029 | 2.115 | 3.785 |
| CXCL3 | 81.438 | 1.572 | 2.468 | 10.269 | 20.713 |
| DKK1 | 2.336 | −4.204 | −2.932 | −1.544 | 1.073 |
| EFNB2 | 1.194 | 1.937 | 1.172 | 2.068 | −1.052 |
| HGF | 1.221 | 2.207 | 1.339 | 1.817 | 1.562 |
| HSD11B1 | 30.422 | 4.501 | 1.147 | 18.48 | 29.309 |
| ID1 | 2.283 | 1.614 | 1.799 | 1.486 | 5.361 |
| IGF1 | 9.016 | 1.147 | 1.068 | 1.793 | 14.374 |
| IGF2 | 1.476 | 3.942 | −1.069 | 5.632 | 4.821 |
| IL1R1 | 3.726 | 2.233 | −1.03 | 2.565 | 3.842 |
| IL6 | 5.484 | 1.272 | 1.155 | 1.505 | 1.657 |
| MDK | 1.975 | 1.966 | 1.07 | 2.106 | 3.017 |
| MEF2C | −2.282 | −1.75 | 1.051 | −2.7 | −2.127 |
| NTN1 | 8.152 | 2.264 | 1.058 | 7.593 | 9.835 |
| OASL | 7.487 | 3.377 | 1.48 | 2.564 | 1.796 |
| PCSK1 | 58.887 | 6.653 | 2.041 | 21.927 | 77.419 |
| PPARG | 1.233 | 2.796 | −1.014 | 2.469 | 2.021 |
| PPARGC1A | 19.933 | 3.017 | 3.13 | 12.553 | 32.307 |
| PPL | 10.936 | 9.128 | 2.676 | 19.03 | 19.77 |
| PTGER4 | −1.016 | 2.245 | 2.538 | 3.785 | 5.318 |
| PTPN22 | 8.385 | 3.023 | 1.19 | 4.239 | 7.19 |
| RAPGEF3 | 2.288 | 2.067 | 1.051 | 2.22 | 4.005 |
| SFRP1 | 6.398 | 6.344 | 2.657 | 6.847 | 6.662 |
| SOD2 | 12.349 | 1.219 | 1.229 | 2.142 | 3.923 |
| STAT1 | 1.141 | −1.865 | −1.121 | −2.134 | −1.723 |
| STC1 | 12.298 | 1.987 | 1.49 | 2.41 | 5.001 |
| TF | 9.482 | 8.842 | 2.214 | 4.156 | 5.078 |
| TNFRSF11B | 2.63 | −2.201 | −4.728 | −1.36 | −1.556 |
| Commonly Upregulated for Control Subject no. 3 Compared to Other Participants in Control Group (25) | Commonly Downregulated for Control Subject no. 3 Compared to Other Participants in Control Group (0) | Downregulated for Control Subject no. 3 and Upregulated for Other Participants in Control Group (3) | Upregulated for Control Subject no. 3 and Downregulated for Other Participants in Control Group (1) |
|---|---|---|---|
| ACKR3 | - | IGF2 | MEF2C |
| BDKRB2 | - | IL1R1 | - |
| C3AR1 | - | PPARG | - |
| CCL5 | - | - | - |
| CFB | - | - | - |
| CSF1 | - | - | - |
| CXCL2 | - | - | - |
| CXCL3 | - | - | - |
| HGF | - | - | - |
| HSD11B1 | - | - | - |
| ID1 | - | - | - |
| IGF1 | - | - | - |
| IL6 | - | - | - |
| MDK | - | - | - |
| NTN1 | - | - | - |
| OASL | - | - | - |
| PCSK1 | - | - | - |
| PPARGC1A | - | - | - |
| PPL | - | - | - |
| PTPN22 | - | - | - |
| RAPGEF3 | - | - | - |
| SFRP1 | - | - | - |
| SOD2 | - | - | - |
| STC1 | - | - | - |
| TF | - | - | - |
| Gene Symbol | no. 1 | no. 2 | no. 3 | no. 4 | no. 5 |
|---|---|---|---|---|---|
| C1QTNF1 | 16.527 | 3.107 | 1.901 | 3.946 | 7.916 |
| CFB | 9.544 | 1.4 | 1.361 | 2.173 | 3.967 |
| CHI3L1 | 15.654 | 2.156 | 1.85 | 2.939 | 16.257 |
| CP | 14.028 | 15.237 | 2.171 | 10.585 | 40.727 |
| CSF1 | 4.442 | 2.414 | 1.685 | 2.024 | 3.503 |
| CXCL2 | 9.746 | 1.278 | 2.029 | 2.115 | 3.785 |
| GNA14 | 8.773 | 4.354 | 1.338 | 7.26 | 47.7 |
| HSD11B1 | 30.422 | 4.501 | 1.147 | 18.48 | 29.309 |
| IGF1 | 9.016 | 1.147 | 1.068 | 1.793 | 14.374 |
| IL6 | 5.484 | 1.272 | 1.155 | 1.505 | 1.657 |
| KCNJ2 | 13.25 | 8.057 | 1.156 | 6.693 | 4.853 |
| KYNU | 21.468 | 5.505 | 3.764 | 17.825 | 35.813 |
| LBP | 47.991 | 98.045 | 3.718 | 66.303 | 84.066 |
| MME | 15.395 | 3.207 | 1.072 | 6.601 | 3.175 |
| NR4A2 | 16.509 | 4.202 | 1.354 | 6.798 | 7.023 |
| NTN1 | 8.152 | 2.264 | 1.058 | 7.593 | 9.835 |
| OAS1 | 8.047 | 3.939 | 2.701 | 3.431 | 7.107 |
| PCSK1 | 58.887 | 6.653 | 2.041 | 21.927 | 77.419 |
| PDK4 | 10.246 | 1.569 | 2.154 | 1.917 | 10.588 |
| PLIN2 | 6.437 | 1.52 | 1.065 | 1.003 | 1.366 |
| PPARGC1A | 19.933 | 3.017 | 3.13 | 12.553 | 32.307 |
| PTGDS | 9.258 | 5.15 | 4.065 | 13.654 | 3.19 |
| SOD2 | 12.349 | 1.219 | 1.229 | 2.142 | 3.923 |
| TF | 9.482 | 8.842 | 2.214 | 4.156 | 5.078 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, Y.; Kim, D.; Choi, Y.J.; Song, I.; Chung, Y.-S. Gene Network Analysis for Osteoporosis, Sarcopenia, Diabetes, and Obesity in Human Mesenchymal Stromal Cells. Genes 2022, 13, 459. https://doi.org/10.3390/genes13030459
Jin Y, Kim D, Choi YJ, Song I, Chung Y-S. Gene Network Analysis for Osteoporosis, Sarcopenia, Diabetes, and Obesity in Human Mesenchymal Stromal Cells. Genes. 2022; 13(3):459. https://doi.org/10.3390/genes13030459
Chicago/Turabian StyleJin, Yilan, Dowan Kim, Yong Jun Choi, Insun Song, and Yoon-Sok Chung. 2022. "Gene Network Analysis for Osteoporosis, Sarcopenia, Diabetes, and Obesity in Human Mesenchymal Stromal Cells" Genes 13, no. 3: 459. https://doi.org/10.3390/genes13030459
APA StyleJin, Y., Kim, D., Choi, Y. J., Song, I., & Chung, Y. -S. (2022). Gene Network Analysis for Osteoporosis, Sarcopenia, Diabetes, and Obesity in Human Mesenchymal Stromal Cells. Genes, 13(3), 459. https://doi.org/10.3390/genes13030459