Figure 1.
Overview of the workflow. TF—transcription factor; IFX—infliximab–methotrexate combination therapy; MTX—methotrexate monotherapy; TCZ—tocilizumab monotherapy.
Figure 1.
Overview of the workflow. TF—transcription factor; IFX—infliximab–methotrexate combination therapy; MTX—methotrexate monotherapy; TCZ—tocilizumab monotherapy.
Figure 2.
Construction of the GTRD-annotated differential co-expression network. The GTRD database contains transcription factors (triangles) and target genes (circles). The presence of a binding site for the transcription factor in the promoter region of the target gene is shown by an arrow pointing from the transcription factor (TF) to the target gene. A TF–TF interaction is also shown. The differential co-expression network has gene pairs that show significantly increased edge weights (purple connector) or significantly decreased edge weights (green connector) in the comparison. The connections are not directed (connector without arrow) and may occur between any two genes. The GTRD-annotated differential co-expression network has TF–TF edges or TF–target gene edges if the edge is present in both GTRD and the differential co-expression network. The direction for the edge is annotated using GTRD. Purple arrows show a TF–target gene or TF–TF interaction where the edge weight is increased, and green arrows show one where the edge weight is decreased. There are no target gene–target gene edges in this network.
Figure 2.
Construction of the GTRD-annotated differential co-expression network. The GTRD database contains transcription factors (triangles) and target genes (circles). The presence of a binding site for the transcription factor in the promoter region of the target gene is shown by an arrow pointing from the transcription factor (TF) to the target gene. A TF–TF interaction is also shown. The differential co-expression network has gene pairs that show significantly increased edge weights (purple connector) or significantly decreased edge weights (green connector) in the comparison. The connections are not directed (connector without arrow) and may occur between any two genes. The GTRD-annotated differential co-expression network has TF–TF edges or TF–target gene edges if the edge is present in both GTRD and the differential co-expression network. The direction for the edge is annotated using GTRD. Purple arrows show a TF–target gene or TF–TF interaction where the edge weight is increased, and green arrows show one where the edge weight is decreased. There are no target gene–target gene edges in this network.
![Genes 13 01216 g002]()
Figure 3.
Number of transcription factor binding sites for transcription factors that interact with PFKFB3, DLD or GAPDH, as annotated by GTRD and GeneHancer. Grey bars represent the number of TF binding sites in the interval −1000 to +100 around the transcription start site of the target gene according to GTRD. Yellow bars represent the number of TF binding sites outside the interval −1000 to +100 around the transcription start site of the target gene according to GeneHancer. (A) The transcription factors binding sites for PFKFB3, (B) The binding sites for DLD, (C) The binding sites for GAPDH.
Figure 3.
Number of transcription factor binding sites for transcription factors that interact with PFKFB3, DLD or GAPDH, as annotated by GTRD and GeneHancer. Grey bars represent the number of TF binding sites in the interval −1000 to +100 around the transcription start site of the target gene according to GTRD. Yellow bars represent the number of TF binding sites outside the interval −1000 to +100 around the transcription start site of the target gene according to GeneHancer. (A) The transcription factors binding sites for PFKFB3, (B) The binding sites for DLD, (C) The binding sites for GAPDH.
Figure 4.
Subgraph of differentially expressed glycolysis-related enzymes with high centrality scores in the GTRD-annotated network of edges with significantly different edge weights between untreated RA and healthy control samples, in CD8+ Tem cells. The differentially expressed transcription factors that link to the enzymes are shown. Triangles indicate transcription factors, and circles indicate target genes. Node colour shows the direction of differential expression: red nodes are over-expressed and blue are under-expressed. The presence of an edge between a TF and a target gene refers to a significant difference in the edge weights of that gene pair between untreated RA samples and healthy samples. Purple edges show higher edge weight in RA, and green shows higher edge weight in healthy samples.
Figure 4.
Subgraph of differentially expressed glycolysis-related enzymes with high centrality scores in the GTRD-annotated network of edges with significantly different edge weights between untreated RA and healthy control samples, in CD8+ Tem cells. The differentially expressed transcription factors that link to the enzymes are shown. Triangles indicate transcription factors, and circles indicate target genes. Node colour shows the direction of differential expression: red nodes are over-expressed and blue are under-expressed. The presence of an edge between a TF and a target gene refers to a significant difference in the edge weights of that gene pair between untreated RA samples and healthy samples. Purple edges show higher edge weight in RA, and green shows higher edge weight in healthy samples.
Figure 5.
Subgraph of differentially expressed glycolysis-related enzymes with high degree centrality in the GeneHancer-annotated network of edges with significantly different edge weights between untreated RA and healthy control samples, in CD8+ Tem cells. The differentially expressed transcription factors that link to the enzymes are shown. Triangles indicate transcription factors and circles indicate target genes. Node colour shows the direction of differential expression: red nodes are over-expressed and blue nodes are under-expressed. The presence of an edge between a TF and a target gene refers to a significant difference in the edge weights of that gene pair between untreated RA samples and healthy samples. Purple edges show higher edge weight in RA, and green shows higher edge weight in healthy samples.
Figure 5.
Subgraph of differentially expressed glycolysis-related enzymes with high degree centrality in the GeneHancer-annotated network of edges with significantly different edge weights between untreated RA and healthy control samples, in CD8+ Tem cells. The differentially expressed transcription factors that link to the enzymes are shown. Triangles indicate transcription factors and circles indicate target genes. Node colour shows the direction of differential expression: red nodes are over-expressed and blue nodes are under-expressed. The presence of an edge between a TF and a target gene refers to a significant difference in the edge weights of that gene pair between untreated RA samples and healthy samples. Purple edges show higher edge weight in RA, and green shows higher edge weight in healthy samples.
Figure 6.
The transcription factors that have edges with both PFKFB3 and PFKFB4 in the RA CD8+ Tem cell GTRD and GeneHancer-annotated networks of differentially co-expressed edges. (A) The subgraph from the GTRD-annotated network (B) The subgraph from the GeneHancer-annotated network. Triangles indicate transcription factors and circles indicate target genes. Node colour shows the direction of differential expression: red nodes are over-expressed and blue nodes are under-expressed. The presence of an edge between a TF and a target gene refers to a significant difference in the edge weights of that gene pair between untreated RA samples and healthy samples. Purple edges show higher edge weight in RA, and green shows higher edge weight in healthy samples.
Figure 6.
The transcription factors that have edges with both PFKFB3 and PFKFB4 in the RA CD8+ Tem cell GTRD and GeneHancer-annotated networks of differentially co-expressed edges. (A) The subgraph from the GTRD-annotated network (B) The subgraph from the GeneHancer-annotated network. Triangles indicate transcription factors and circles indicate target genes. Node colour shows the direction of differential expression: red nodes are over-expressed and blue nodes are under-expressed. The presence of an edge between a TF and a target gene refers to a significant difference in the edge weights of that gene pair between untreated RA samples and healthy samples. Purple edges show higher edge weight in RA, and green shows higher edge weight in healthy samples.
Figure 7.
Subgraph of differentially expressed glycolysis-related enzymes with high centrality scores in the GTRD-annotated network of edges with significantly different edge weights between tocilizumab-treated RA and untreated RA samples, in CD8+ Tem cells. The differentially expressed transcription factors that link to the enzymes are shown. Triangles indicate transcription factors, and circles indicate target genes. Node colour shows the direction of differential expression: red nodes are over-expressed and blue nodes are under-expressed. The presence of an edge between a TF and a target gene refers to a significant difference in the edge weights of that gene pair between untreated RA samples and healthy samples. Purple edges show higher edge weight in RA, and green shows higher edge weight in healthy samples.
Figure 7.
Subgraph of differentially expressed glycolysis-related enzymes with high centrality scores in the GTRD-annotated network of edges with significantly different edge weights between tocilizumab-treated RA and untreated RA samples, in CD8+ Tem cells. The differentially expressed transcription factors that link to the enzymes are shown. Triangles indicate transcription factors, and circles indicate target genes. Node colour shows the direction of differential expression: red nodes are over-expressed and blue nodes are under-expressed. The presence of an edge between a TF and a target gene refers to a significant difference in the edge weights of that gene pair between untreated RA samples and healthy samples. Purple edges show higher edge weight in RA, and green shows higher edge weight in healthy samples.
Figure 8.
Subgraph of MYC, PPARG and INSR and their target genes in the GTRD-annotated network of edges with significantly different edge weights between untreated RA and healthy samples in CD8+ Temra cells. The differentially expressed transcription factors that link to the enzymes are shown. Triangles indicate transcription factors and circles indicate target genes. Node colour shows the direction of differential expression: red nodes are over-expressed, blue nodes are under-expressed and white nodes are not differentially expressed. The presence of an edge between a TF and a target gene refers to a significant difference in the edge weights of that gene pair between untreated RA samples and healthy samples. Purple edges show higher edge weight in RA, and green shows a higher edge weight in healthy samples.
Figure 8.
Subgraph of MYC, PPARG and INSR and their target genes in the GTRD-annotated network of edges with significantly different edge weights between untreated RA and healthy samples in CD8+ Temra cells. The differentially expressed transcription factors that link to the enzymes are shown. Triangles indicate transcription factors and circles indicate target genes. Node colour shows the direction of differential expression: red nodes are over-expressed, blue nodes are under-expressed and white nodes are not differentially expressed. The presence of an edge between a TF and a target gene refers to a significant difference in the edge weights of that gene pair between untreated RA samples and healthy samples. Purple edges show higher edge weight in RA, and green shows a higher edge weight in healthy samples.
Figure 9.
Subgraph of MYC, PPARG and INSR and their target genes in the GeneHancer annotated network of edges with significantly different edge weights between untreated RA and healthy samples, in CD8+ Temra cells. The differentially expressed transcription factors that link to the enzymes are shown. Triangles indicate transcription factors and circles indicate target genes. Node color shows the direction of differential expression: red nodes are over-expressed, blue nodes are under-expressed and white nodes are not differentially expressed. The presence of an edge between a TF and a target gene refers to a significant difference in the edge weights of that gene pair between untreated RA samples and healthy samples. Purple edges show higher edge weight in RA, and green shows higher edge weight in healthy samples.
Figure 9.
Subgraph of MYC, PPARG and INSR and their target genes in the GeneHancer annotated network of edges with significantly different edge weights between untreated RA and healthy samples, in CD8+ Temra cells. The differentially expressed transcription factors that link to the enzymes are shown. Triangles indicate transcription factors and circles indicate target genes. Node color shows the direction of differential expression: red nodes are over-expressed, blue nodes are under-expressed and white nodes are not differentially expressed. The presence of an edge between a TF and a target gene refers to a significant difference in the edge weights of that gene pair between untreated RA samples and healthy samples. Purple edges show higher edge weight in RA, and green shows higher edge weight in healthy samples.
Figure 10.
Normalized counts of genes associated with metabolic pathways or the regulation of glycolysis that show differential regulation in untreated RA CD8+ Tem cells or in TCZ-treated RA CD8+ Tem cells. Significant differential regulation is shown by asterisks (***).
Figure 10.
Normalized counts of genes associated with metabolic pathways or the regulation of glycolysis that show differential regulation in untreated RA CD8+ Tem cells or in TCZ-treated RA CD8+ Tem cells. Significant differential regulation is shown by asterisks (***).
Figure 11.
Normalized counts of genes associated with metabolic pathways or the regulation of glycolysis that show differential regulation in untreated RA CD8+ Tem cells or in untreated RA CD8+ Temra cells. Significant differential regulation is shown by asterisks (***).
Figure 11.
Normalized counts of genes associated with metabolic pathways or the regulation of glycolysis that show differential regulation in untreated RA CD8+ Tem cells or in untreated RA CD8+ Temra cells. Significant differential regulation is shown by asterisks (***).
Figure 12.
The differentially expressed genes of RA CD8+ Tem cells in the PI3K-AKT pathway. Red indicates upregulated genes and blue indicates downregulated genes.
Figure 12.
The differentially expressed genes of RA CD8+ Tem cells in the PI3K-AKT pathway. Red indicates upregulated genes and blue indicates downregulated genes.
Figure 13.
The differentially expressed genes of RA CD8+ Tem cells in the mTOR signalling pathway. Red indicates upregulated genes and blue indicates downregulated genes.
Figure 13.
The differentially expressed genes of RA CD8+ Tem cells in the mTOR signalling pathway. Red indicates upregulated genes and blue indicates downregulated genes.
Figure 14.
The differentially expressed genes of RA CD8+ Temra cells in the PI3K-AKT pathway. Red indicates upregulated genes and blue indicates downregulated genes.
Figure 14.
The differentially expressed genes of RA CD8+ Temra cells in the PI3K-AKT pathway. Red indicates upregulated genes and blue indicates downregulated genes.
Figure 15.
The differentially expressed genes of RA CD8+ Temra cells in the mTOR pathway. Red indicates upregulated genes and blue indicates downregulated genes.
Figure 15.
The differentially expressed genes of RA CD8+ Temra cells in the mTOR pathway. Red indicates upregulated genes and blue indicates downregulated genes.
Figure 16.
Differential expression of enzymes involved in glycolysis, pentose phosphate pathway, TCA cycle and oxidative phosphorylation in RA CD8+ Tem cells. Solid red UP arrows indicate upregulation/increased activity. Solid blue down arrows indicate downregulation. Light red arrows indicate statistically significant upregulation with low read counts. Pathways where the overall effect of differential regulation on the activity of the pathway is ambiguous are marked by a question mark. PPP—pentose phosphate pathway; G6P—glucose-6-phosphate; F6P—fructose-6-phosphate; GA3P—glyceraldehyde-3-phosphate.
Figure 16.
Differential expression of enzymes involved in glycolysis, pentose phosphate pathway, TCA cycle and oxidative phosphorylation in RA CD8+ Tem cells. Solid red UP arrows indicate upregulation/increased activity. Solid blue down arrows indicate downregulation. Light red arrows indicate statistically significant upregulation with low read counts. Pathways where the overall effect of differential regulation on the activity of the pathway is ambiguous are marked by a question mark. PPP—pentose phosphate pathway; G6P—glucose-6-phosphate; F6P—fructose-6-phosphate; GA3P—glyceraldehyde-3-phosphate.
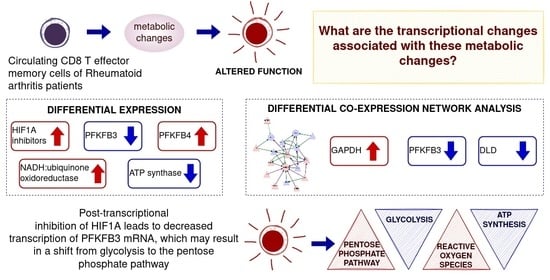
Figure 17.
Proposed model for the regulation of glycolysis and pentose phosphate pathway in RA CD8+ Tem cells. EGLN3, ELOB and GAPDH are upregulated as indicated by the red up arrows. This leads to the inhibition of HIF1A (red T—shaped connector). HIF1A-mediated upregulation of PFKFB3 (black dotted arrow) is blocked (red X) leading to the downregulation of PFKFB3 (blue down arrow). The PFKFB3 catalysed increase in fructose 2, 6 bisphosphate (F 2,6 BP) is abolished, so F 2,6 BP-mediated allosteric activation of phosphofructokinase (PFK) is lost, and the commitment step of glycolysis does not occur. This leads to reduced glycolysis. PFKFB3 downregulation causes a reduction in the ratio of PFKFB3 with G6PD (glucose 6 phosphate dehydrogenase) leading to increased pentose phosphate pathway activity.
Figure 17.
Proposed model for the regulation of glycolysis and pentose phosphate pathway in RA CD8+ Tem cells. EGLN3, ELOB and GAPDH are upregulated as indicated by the red up arrows. This leads to the inhibition of HIF1A (red T—shaped connector). HIF1A-mediated upregulation of PFKFB3 (black dotted arrow) is blocked (red X) leading to the downregulation of PFKFB3 (blue down arrow). The PFKFB3 catalysed increase in fructose 2, 6 bisphosphate (F 2,6 BP) is abolished, so F 2,6 BP-mediated allosteric activation of phosphofructokinase (PFK) is lost, and the commitment step of glycolysis does not occur. This leads to reduced glycolysis. PFKFB3 downregulation causes a reduction in the ratio of PFKFB3 with G6PD (glucose 6 phosphate dehydrogenase) leading to increased pentose phosphate pathway activity.
Figure 18.
Differentially expressed transcription factors which show differential co-expression with GAPDH in RA CD8+ Tem cells. Red boxes show genes that are upregulated, and blue boxes show those that are downregulated. The transcription factors with increased edge weights are shown in the purple box and those with reduced edge weights with GAPDH in RA are shown in the green box. The functions of each transcription factor are also shown.
Figure 18.
Differentially expressed transcription factors which show differential co-expression with GAPDH in RA CD8+ Tem cells. Red boxes show genes that are upregulated, and blue boxes show those that are downregulated. The transcription factors with increased edge weights are shown in the purple box and those with reduced edge weights with GAPDH in RA are shown in the green box. The functions of each transcription factor are also shown.
Figure 19.
RA CD8+ Tem differentially expressed genes in the PI3K-AKT pathway and the mTOR pathway. Red up arrows indicate upregulation and blue down arrows indicate downregulation. Red connectors indicate the effect of the differentially regulated genes on their targets, whereas T shaped arrows indicate the inhibition and normal arrows indicate activation.
Figure 19.
RA CD8+ Tem differentially expressed genes in the PI3K-AKT pathway and the mTOR pathway. Red up arrows indicate upregulation and blue down arrows indicate downregulation. Red connectors indicate the effect of the differentially regulated genes on their targets, whereas T shaped arrows indicate the inhibition and normal arrows indicate activation.
Figure 20.
The differentially regulated genes involved in the regulation of glycolysis in RA CD8+ Tem cells. Post transcriptional inhibition of HIF1A by the upregulated ELOB, EGLN3 and GAPDH (red up arrow) inhibits HIF1A activity resulting in the downregulation of PFKFB3 (blue down arrow). This causes reduced activity of the glycolytic enzyme PFK (light blue down arrow) and increased activity of the pentose phosphate pathway enzyme G6PD (light red up arrow). The upregulated transcription factor TP63 (red up arrow) causes increased expression of GAPDH (red up arrow). IRS2 is downregulated (blue down arrow), while RPS6KA1, HRAS and LAMTOR2 are upregulated (red up arrow) upstream of mTOR. The effect of these changes in mTOR cannot be determined in this study (question mark). The pentose phosphate pathway genes, IDNK and TALDO1, are also upregulated (red up arrow).
Figure 20.
The differentially regulated genes involved in the regulation of glycolysis in RA CD8+ Tem cells. Post transcriptional inhibition of HIF1A by the upregulated ELOB, EGLN3 and GAPDH (red up arrow) inhibits HIF1A activity resulting in the downregulation of PFKFB3 (blue down arrow). This causes reduced activity of the glycolytic enzyme PFK (light blue down arrow) and increased activity of the pentose phosphate pathway enzyme G6PD (light red up arrow). The upregulated transcription factor TP63 (red up arrow) causes increased expression of GAPDH (red up arrow). IRS2 is downregulated (blue down arrow), while RPS6KA1, HRAS and LAMTOR2 are upregulated (red up arrow) upstream of mTOR. The effect of these changes in mTOR cannot be determined in this study (question mark). The pentose phosphate pathway genes, IDNK and TALDO1, are also upregulated (red up arrow).
Figure 21.
RA CD8+ Temra differentially expressed genes in the PI3K-AKT pathway and the mTOR pathway. Red up arrows indicate upregulation. Red arrows indicate the effect of the differentially regulated genes on their targets, where T shaped arrows indicate inhibition and normal arrows indicate activation. Dotted arrows indicate the presence of intermediate steps between the two connected proteins. Red crosses show the interactions that are lost in the RA CD8+ Temra cells due to inhibition or downregulation of the upstream proteins.
Figure 21.
RA CD8+ Temra differentially expressed genes in the PI3K-AKT pathway and the mTOR pathway. Red up arrows indicate upregulation. Red arrows indicate the effect of the differentially regulated genes on their targets, where T shaped arrows indicate inhibition and normal arrows indicate activation. Dotted arrows indicate the presence of intermediate steps between the two connected proteins. Red crosses show the interactions that are lost in the RA CD8+ Temra cells due to inhibition or downregulation of the upstream proteins.
Figure 22.
The differentially regulated genes involved in the regulation of glycolysis in RA CD8+ Temra cells. Post transcriptional inhibition of HIF1A by the upregulated ELOB and EGLN3 (red up arrow) and the downregulation of HIF1A co-activator EP300 (blue down arrow) inhibit HIF1A activity. AKT1S1 is downregulated (blue down arrow), while RPS6KA1, IL6R, WNT7A and LRP5 are upregulated (red up arrow) upstream of mTOR. AKT and mTOR may be activated in RA CD8+ Temra cells (light red up arrow and question mark). This may be the cause of the upregulation of MYC (red up arrow). Glycolysis target genes of MYC and HIF1A do not show differential regulation (black = signs). AKT and mTOR activity may increase the activity of glycolysis genes HK, PDK1, PFKFB2 and GLUT1-4 (light red up arrow and question mark). ENO3 and PDHA1 are upregulated (red up arrow).
Figure 22.
The differentially regulated genes involved in the regulation of glycolysis in RA CD8+ Temra cells. Post transcriptional inhibition of HIF1A by the upregulated ELOB and EGLN3 (red up arrow) and the downregulation of HIF1A co-activator EP300 (blue down arrow) inhibit HIF1A activity. AKT1S1 is downregulated (blue down arrow), while RPS6KA1, IL6R, WNT7A and LRP5 are upregulated (red up arrow) upstream of mTOR. AKT and mTOR may be activated in RA CD8+ Temra cells (light red up arrow and question mark). This may be the cause of the upregulation of MYC (red up arrow). Glycolysis target genes of MYC and HIF1A do not show differential regulation (black = signs). AKT and mTOR activity may increase the activity of glycolysis genes HK, PDK1, PFKFB2 and GLUT1-4 (light red up arrow and question mark). ENO3 and PDHA1 are upregulated (red up arrow).
Table 1.
Details of the samples in the data set GSE118829.
Table 1.
Details of the samples in the data set GSE118829.
| Group | Cell Type | Total Samples | Gender | Age |
|---|
| Healthy control | CD4+ Tn | 9 | 7f, 2m | 45–77 |
| | CD4+ Tcm | 10 | 8f, 2m | |
| | CD4+ Tem | 10 | 8f, 2m | |
| | CD8+ Tn | 10 | 8f, 2m | |
| | CD8+ Tcm | 8 | 6f, 2m | |
| | CD8+ Tem | 10 | 8f, 2m | |
| | CD8+ Temra | 9 | 7f, 2m | |
| Untreated RA | CD4+ Tn | 10 | 8f, 2m | 46–68 |
| | CD4+ Tcm | 10 | 8f, 2m | |
| | CD4+ Tem | 9 | 7f, 2m | |
| | CD8+ Tn | 9 | 7f, 2m | |
| | CD8+ Tcm | 7 | 5f, 2m | |
| | CD8+ Tem | 8 | 7f, 1m | |
| | CD8+ Temra | 8 | 6f, 2m | |
| Infliximab-treated | CD4+ Tn | 9 | 8f, 1m | 44–66 |
| | CD4+ Tcm | 10 | 9f, 1m | |
| | CD4+ Tem | 9 | 8f, 1m | |
| | CD8+ Tn | 9 | 8f, 1m | |
| | CD8+ Tcm | 10 | 9f, 1m | |
| | CD8+ Tem | 6 | 5f, 1m | |
| | CD8+ Temra | 8 | 7f, 1m | |
| Methotrexate-treated | CD4+ Tn | 10 | 9f, 1m | 28–76 |
| | CD4+ Tcm | 9 | 8f, 1m | |
| | CD4+ Tem | 8 | 7f, 1m | |
| | CD8+ Tn | 8 | 7f, 1m | |
| | CD8+ Tcm | 4 | 3f, 1m | |
| | CD8+ Tem | 9 | 8f, 1m | |
| | CD8+ Temra | 6 | 5f, 1m | |
| Tocilizumab-treated | CD4+ Tn | 10 | 10f | 44–73 |
| | CD4+ Tcm | 10 | 10f | |
| | CD4+ Tem | 10 | 10f | |
| | CD8+ Tn | 10 | 10f | |
| | CD8+ Tcm | 8 | 8f | |
| | CD8+ Tem | 10 | 10f | |
| | CD8+ Temra | 7 | 7f | |
| Synovial fluid | CD4+ Tn | 0 | | 71 |
| | CD4+ Tcm | 4 | 3f, 1m | |
| | CD4+ Tem | 3 | 3f | |
| | CD8+ Tn | 0 | | |
| | CD8+ Tcm | 1 | 1m | |
| | CD8+ Tem | 1 | 1f | |
| | CD8+ Temra | 1 | 1f | |
Table 2.
Upregulated and downregulated genes of untreated RA compared with healthy samples of T cell subsets.
Table 2.
Upregulated and downregulated genes of untreated RA compared with healthy samples of T cell subsets.
| Cell Type | Upregulated Genes | Downregulated Genes |
|---|
| CD4+ Tn | 12 | 5 |
| CD4+ Tcm | 10 | 63 |
| CD4+ Tem | 3 | 1 |
| CD8+ Tn | 21 | 5 |
| CD8+ Tcm | 3 | 7 |
| CD8+ Tem | 1617 | 164 |
| CD8+ Temra | 843 | 291 |
Table 3.
Upregulated and downregulated genes of treated RA compared with untreated RA samples of T cell subsets.
Table 3.
Upregulated and downregulated genes of treated RA compared with untreated RA samples of T cell subsets.
| Cell Type | Methotrexate | Infliximab | Tocilizumab |
|---|
| UP | DOWN | UP | DOWN | UP | DOWN |
|---|
| CD4+ Tn | 1 | 9 | 12 | 4 | 4 | 20 |
| CD4+ Tcm | 1 | 1 | 1 | 8 | 2 | 4 |
| CD4+ Tem | 2 | 2 | 0 | 3 | 4 | 6 |
| CD8+ Tn | 16 | 51 | 0 | 26 | 19 | 711 |
| CD8+ Tcm | 3 | 6 | 2 | 3 | 3 | 3 |
| CD8+ Tem | 26 | 13 | 4 | 7 | 91 | 1047 |
| CD8+ Temra | 45 | 266 | 35 | 130 | 12 | 59 |
Table 4.
Enriched transcription factor of the downregulated gene list from untreated RA CD8+ Tem cells.
Table 4.
Enriched transcription factor of the downregulated gene list from untreated RA CD8+ Tem cells.
| Term | Overlap | p-Value | Adjusted p-Value | Odds Ratio |
|---|
| CREB1 CHEA | 30/1444 | 2.04 × 10−6 | 0.0001 | 2.92 |
| UBTF ENCODE | 33/1631 | 1.02 × 10−6 | 0.0001 | 2.88 |
| CREB1 ENCODE | 38/2238 | 9.19 × 10−6 | 0.0003 | 2.42 |
| ZNF384 ENCODE | 17/730 | 0.00011 | 0.0026 | 3.10 |
| SUZ12 CHEA | 25/1684 | 0.0027 | 0.0525 | 1.97 |
| PBX3 ENCODE | 20/1269 | 0.0038 | 0.0616 | 2.07 |
| ATF2 ENCODE | 36/2852 | 0.005 | 0.0689 | 1.69 |
| BHLHE40 ENCODE | 8/348 | 0.0081 | 0.0996 | 2.94 |
| ZEB1 ENCODE | 4/106 | 0.011 | 0.1010 | 4.84 |
| MAX ENCODE | 27/2073 | 0.010 | 0.1010 | 1.71 |
Table 5.
Enriched transcription factor of the downregulated gene list from untreated RA CD8+ Temra cells.
Table 5.
Enriched transcription factor of the downregulated gene list from untreated RA CD8+ Temra cells.
| Term | Overlap | p-Value | Adjusted p-Value | Odds Ratio |
|---|
| TAF1 ENCODE | 95/3346 | 1.69 × 10−11 | 1.73 × 10−9 | 2.45 |
| CREB1 CHEA | 51/1444 | 2.89 × 10−9 | 1.48 × 10−7 | 2.79 |
| GABPA ENCODE | 64/2082 | 4.99 × 10−9 | 1.69 × 10−7 | 2.47 |
| UBTF ENCODE | 53/1631 | 2.30 × 10−8 | 5.87 × 10−7 | 2.56 |
| CREB1 ENCODE | 65/2238 | 3.34 × 10−8 | 6.82 × 10−7 | 2.32 |
| BRCA1 ENCODE | 82/3218 | 1.14 × 10−7 | 1.95 × 10−6 | 2.073 |
| ELF1 ENCODE | 68/2483 | 1.45 × 10−7 | 2.11 × 10−6 | 2.18 |
| ZNF384 ENCODE | 30/730 | 3.34 × 10−7 | 4.25 × 10−6 | 3.12 |
| ATF2 ENCODE | 73/2852 | 6.29 × 10−7 | 7.13 × 10−6 | 2.04 |
| YY1 ENCODE | 70/2753 | 1.48 × 10−6 | 1.51 × 10−5 | 2.01 |
Table 6.
Glycolysis/gluconeogenesis pathway DEGs in untreated RA T cell subsets. Means are calculated from DESeq2 normalized counts.
Table 6.
Glycolysis/gluconeogenesis pathway DEGs in untreated RA T cell subsets. Means are calculated from DESeq2 normalized counts.
| Cell Type | Gene | adj. Pval | Log2FC | Linear FC | RA Mean | Control Mean |
|---|
| CD8+ Tem | DLD | 0.071 | −0.664 | 0.63 | 126.84 | 203.29 |
| CD8+ Tem | GAPDH | 0.052 | 0.631 | 1.55 | 12,014.37 | 7766.11 |
| CD8+ Temra | DLD | 0.046 | −0.802 | 0.57 | 96.47 | 169.54 |
| CD8+ Temra | ENO3 | 0.062 | 0.743 | 1.67 | 195.64 | 117.34 |
| CD8+ Temra | PDHA1 | 0.00016 | 0.812 | 1.76 | 220.72 | 125.34 |
| CD8+ Temra | HKDC1 | 0.017 | 2.414 | 5.33 | 6.095 | 1.14 |
| CD8+ Temra | HK2 | 0.004 | 1.894 | 3.72 | 9.49 | 2.58 |
Table 7.
Glycolysis/gluconeogenesis pathway DEGs in MTX-, IFX- and TCZ-treated RA T cell subsets. Means are calculated from DESeq2 normalized counts.
Table 7.
Glycolysis/gluconeogenesis pathway DEGs in MTX-, IFX- and TCZ-treated RA T cell subsets. Means are calculated from DESeq2 normalized counts.
| Treatment | Cell Type | Gene | adj. Pval | Log2FC | Linear FC | Treatment Mean | RA Mean |
|---|
| TCZ | CD4+ Tn | LDHA | 0.0008 | −0.74 | 0.597 | 572.38 | 947.52 |
| TCZ | CD8+ Tem | PCK2 | 0.097 | 0.67 | 1.56 | 82.23 | 52.53 |
| TCZ | CD8+ Tem | GAPDH | 0.055 | −0.66 | 0.63 | 7657.78 | 12,014.37 |
| MTX | CD8+ Temra | HK2 | 0.003 | −3.01 | 0.12 | 1.23 | 9.49 |
Table 8.
Tricarboxylic acid cycle DEGs in untreated RA T cell subsets. Means are calculated from DESeq2 normalized counts.
Table 8.
Tricarboxylic acid cycle DEGs in untreated RA T cell subsets. Means are calculated from DESeq2 normalized counts.
| Cell Type | Gene | adj. Pval | Log2FC | Linear FC | RA Mean | Control Mean |
|---|
| CD8+ Tem | DLD | 0.07 | −0.66 | 0.63 | 126.84 | 203.29 |
| CD8+ Tem | OGDH | 0.088 | 0.67 | 1.59 | 1924.58 | 1212.56 |
| CD8+ Tem | IDH3A | 0.045 | 1.7 | 3.25 | 8.13 | 2.08 |
| CD8+ Temra | DLD | 0.046 | −0.802 | 0.574 | 96.47 | 169.54 |
| CD8+ Temra | PDHA1 | 0.00016 | 0.81 | 1.76 | 220.72 | 125.34 |
Table 9.
Oxidative phosphorylation DEGs in untreated RA CD8+ Tem cells. Means are calculated from DESeq2 normalized counts.
Table 9.
Oxidative phosphorylation DEGs in untreated RA CD8+ Tem cells. Means are calculated from DESeq2 normalized counts.
| Gene | adj. Pval | Log2FC | Linear FC | RA Mean | Control Mean |
|---|
| COX6A1 | 0.095 | 1.068 | 2.097 | 8.67 | 3.86 |
| NDUFB10 | 0.074 | 1.076 | 2.109 | 247.30 | 115.93 |
| NDUFB11 | 0.054 | 1.126 | 2.182 | 721.67 | 327.37 |
| ATP6V1G3 | 0.051 | 5.091 | 34.072 | 7.59 | 0.06 |
| NDUFS2 | 0.077 | 0.691 | 1.615 | 380.8 | 234.79 |
| ATP5F1C | 0.093 | −0.89 | 0.539 | 225.44 | 425.32 |
| ATP5MG | 0.05 | 1.597 | 3.026 | 6.82 | 1.89 |
| COX8A | 0.018 | 1.685 | 3.214 | 20.92 | 6.11 |
| NDUFA12 | 0.042 | 0.602 | 1.518 | 393.41 | 256.95 |
| MT-CO1 | 0.036 | 2.88 | 7.36 | 6.4 | 0.86 |
| UQCRHL | 0.017 | 1.331 | 2.517 | 42.81 | 16.45 |
Table 10.
Oxidative phosphorylation DEGs in TCZ-treated RA T cell subsets. Means are calculated from DESeq2 normalized counts.
Table 10.
Oxidative phosphorylation DEGs in TCZ-treated RA T cell subsets. Means are calculated from DESeq2 normalized counts.
| Cell Type | Gene | adj. Pval | Log2FC | Linear FC | Treatment Mean | RA Mean |
|---|
| CD8+ Tem | ATP5MG | 0.059 | −1.652 | 0.318 | 1.77 | 6.82 |
| CD8+ Tem | COX8A | 0.088 | −1.334 | 0.397 | 8.03 | 20.93 |
| CD8+ Tn | NDUFB6 | 0.088 | −1.541 | 0.344 | 1.94 | 5.88 |
| CD8+ Tn | NDUFB8 | 0.005 | −2.297 | 0.203 | 3.2 | 17.97 |
| CD8+ Tn | MT-CYB | 0.066 | −3.058 | 0.120 | 0.92 | 8.42 |
| CD8+ Tn | MT-ND5 | 0.067 | −2.555 | 0.170 | 2.48 | 14.81 |
| CD8+ Tn | MT-CO1 | 0.071 | −2.757 | 0.148 | 1.00 | 7.01 |
Table 11.
Fatty acid metabolism DEGs in untreated RA T cell subsets. Means are calculated from DESeq2 normalized counts.
Table 11.
Fatty acid metabolism DEGs in untreated RA T cell subsets. Means are calculated from DESeq2 normalized counts.
| Cell Type | Gene | adj. Pval | Log2FC | Linear FC | RA Mean | Control Mean |
|---|
| CD8+ Tem | ACAA2 | 0.099 | 3.165 | 8.972 | 6.4 | 0.65 |
| CD8+ Temra | PECR | 0.028 | 0.666 | 1.587 | 85.27 | 53.35 |
| CD8+ Temra | ACOT1 | 1.0196 × 10−5 | 2.355 | 5.115 | 35.82 | 6.09 |
Table 12.
Fatty acid metabolism DEGs in treated RA T cell subsets. Means are calculated from DESeq2 normalized counts.
Table 12.
Fatty acid metabolism DEGs in treated RA T cell subsets. Means are calculated from DESeq2 normalized counts.
| Treatment | Cell Type | Gene | adj. Pval | Log2FC | Linear FC | Treatment Mean | RA Mean |
|---|
| IFX | CD8+ Temra | ACOT1 | 6.64 × 10−5 | −2.444 | 0.184 | 5.66 | 35.82 |
| TCZ | CD8+ Tem | ACACB | 0.022 | 0.726 | 1.654 | 145.99 | 89.00 |
| TCZ | CD8+ Temra | ACOT1 | 0.0014 | −2.407 | 0.189 | 5.83 | 35.82 |
| TCZ | CD8+ Temra | ACOT2 | 0.029 | −0.832 | 0.562 | 54.80 | 101.98 |
Table 13.
Number of significant edges and GTRD-annotated significant edges in T cell subsets.
Table 13.
Number of significant edges and GTRD-annotated significant edges in T cell subsets.
| Cell Type | RA Untreated vs. Healthy | IFX Treated vs. RA Untreated | MTX Treated vs. RA Untreated | TCZ Treated vs. RA Untreated |
|---|
| Total No: Significant Edges | Significant Edges Present in GTRD | Total No: Significant Edges | Significant Edges Present in GTRD | Total No: Significant Edges | Significant Edges Present in GTRD | Total No: Significant Edges | Significant Edges Present in GTRD |
|---|
| CD4+ Tn | 185 | 127 | 2704 | 1160 | 41 | 24 | 404 | 196 |
| CD4+ Tcm | 15 | 5 | 0 | 0 | 1 | 0 | 753 | 257 |
| CD4+ Tem | 2039 | 1244 | 2521 | 1767 | 1551 | 978 | 6308 | 4205 |
| CD8+ Tn | 22,834 | 12,329 | 11,596 | 6475 | 19,241 | 12,321 | 15,036 | 8528 |
| CD8+ Tcm | 17,414 | 7874 | 331 | 234 | 1929 | 1470 | 38 | 32 |
| CD8+ Tem | 27,355 | 15,411 | 13,678 | 8197 | 28,653 | 15,138 | 17,151 | 9149 |
| CD8+ Temra | 128,318 | 75,606 | 96,729 | 53,945 | 84,906 | 48,879 | 82,597 | 45,443 |
Table 14.
Number of nodes and edges in the GTRD-annotated transcription factor–target gene network of untreated RA and treated RA T cell subsets.
Table 14.
Number of nodes and edges in the GTRD-annotated transcription factor–target gene network of untreated RA and treated RA T cell subsets.
| Cell Type | RA Untreated vs. Healthy | IFX Treated vs. RA Untreated | MTX Treated vs. RA Untreated | TCZ Treated vs. RA Untreated |
|---|
| | Total Nodes, TFs, Target Genes | Total Edges | Total Nodes, TFs, Target Genes | Total Edges | Total Nodes, TFs, Target Genes | Total Edges | Total Nodes, TFs, Target Genes | Total Edges |
|---|
| CD4+ Tn | 92, 88, 4 | 127 | 650, 624, 26 | 1160 | 23, 23, 0 | 24 | 131, 123, 8 | 196 |
| CD4+ Tcm | 7, 7, 0 | 5 | 0 | 0 | 0 | 0 | 207, 202, 5 | 257 |
| CD4+ Tem | 627, 592, 35 | 1244 | 766, 728, 38 | 1767 | 588, 560, 28 | 978 | 1035, 984, 51 | 4205 |
| CD8+ Tn | 1173, 1115, 58 | 12,329 | 1074, 1020, 54 | 6475 | 1136, 1076, 60 | 12,321 | 1183, 1123, 60 | 8528 |
| CD8+ Tcm | 1072, 1018, 54 | 7874 | 182, 176, 6 | 234 | 438, 420, 18 | 1470 | 31, 31, 0 | 32 |
| CD8+ Tem | 1211, 1149, 62 | 15,411 | 1136, 1077, 59 | 8197 | 1248, 1183, 65 | 15,138 | 1146, 1092, 54 | 9149 |
| CD8+ Temra | 1285, 1214, 71 | 75,606 | 1279, 1211, 68 | 53,945 | 1281, 1211, 70 | 48,879 | 1284, 1215, 69 | 45,443 |
Table 15.
Top transcription factors and target genes in GTRD-annotated transcription factor–target gene network of untreated RA and treated RA T cell subsets.
Table 15.
Top transcription factors and target genes in GTRD-annotated transcription factor–target gene network of untreated RA and treated RA T cell subsets.
| Cell Type | RA Untreated v Healthy | IFX-treated v RA Untreated | MTX Treated v RA Untreated | TCZ Treated v RA Untreated |
|---|
| Top Ten TF | Top Ten Target Genes | Top Ten TF | Top Ten Target Genes | Top Ten TF | Top Ten Target Genes | Top Ten TF | Top Ten Target Genes |
|---|
| CD4+ Tn | - | - | FOXA1, HOXA6, MITF, SMAD5, DNMT3B, RAD21, GATAD2B | PFKP, DLD, GAPDHS, PDK1, PGM2, ACSS1 | - | - | - | - |
| CD4+ Tcm | - | - | - | - | - | - | - | - |
| CD4+ Tem | SPI1, PADI2, ZNF366, TP63, EHF, TWIST1, CBFA2T3, PRDM6 | PDK4, GAPDH, PFKP, HK1, ALDH3B1, PGAM1 | SPI1, PADI2, EHF, ZNF366, CBFA2T3, TP63, TWIST1, PPARG, VDR | ALDH3B2, PFKP, PGM2, ADPGK, ADH5 | PADI2, ZNF366, SPI1, TP63, TWIST1, CBFA2T3, EHF | ENO3, PDK4, PGM1, GCK | PADI2, CBFA2T3, ETV1, SPI1, RBBP4, ZNF366, NR2F2, NFE2 | ALDH3B2, ENO3, DLAT, PGM1, ACLY |
| CD8+ Tn | AHR, SALL3, FOXP1, KLF1, L3MBTL2 | ALDH3B1, PCK1, ACSS2, KHK, ALDH7A1, PKM, ALDH3B2, AKR1A1, ENO3 | MYOD1, AHR, RAD21, JUN, FOXP1, L3MBTL2 | ALDH3B1, PCK1, ALDH7A1, KHK, ENO3, ACSS2, AKR1A1, GPI | NELFA, FOXP1, PHF8, HIF1A, RAD21, RING1 | GPI, ALDH3B1, PCK1, PFKL, ADPGK, LDHA, ENO3, ACSS2, PFKFB3 | TAL1, AHR, FOXP1, NR5A2 | ALDH3B1, ALDH7A1, PCK1, KHK, ACSS2, PDK4, ENO3, AKR1A1 |
| CD8+ Tcm | TEAD4, EGR2, SMC1A, FEZF1 | PFKM, PDK2, ALDH3A1, ALDH3B1, PFKL, ALDH3A2, HK3, ADPGK | - | - | - | - | - | - |
| CD8+ Tem | MBL2, E2F8, ME1, ATF3, E2F7 | PFKFB3, GAPDH, BPGM, DLD, G6PD, SLC2A1, PDHA1, PGAM1 | E2F8, ME1, MBL2, E2F7, BRCA1, RBBP4, ATF3, GLIS1, CEBPD | GAPDH, ENO1, PGM1, PGK1, DLD, ENO3, PKM | MBL2, FOXP1, TCF7L1, EGR1, EGR2, ATF3 | ENO2, ALDH2, GAPDH, ALDH9A1, ENO1, PGAM1, GCK | MBL2, E2F8, SMAD1, ME1 | GAPDH, G6PD, ENO2, SLC2A1, G6PC, BPGM, PCK2 |
| CD8+ Temra | MYC, ZNF143, TP53BP1, SP5, CDK7 | ENO1, G6PC3, ALDOC, SLC2A1, ENO3, ACSS2, HK2, AKR1A1, HK1 | ZNF143, TP53BP1, TEAD1, CREB1 | ALDOC, ACSS2, ENO1, HK2, ENO3, G6PD, G6PC3, PFKFB2, AKR1A1, PGM2 | RUNX1T1, MYC, ZNF143, INTS12, TEAD1 | ALDOA, AKR1A1, ACSS2, ENO1, HK2, ALDOC, G6PC3, KHK | BRD3, TP53BP1, TEAD1, GRHL2, ZNF143, INTS12 | ALDOA, ALDOC, G6PC3, ACSS2, ENO1, ENO3, HK2, PFKFB2 |
Table 16.
Top ten target genes in GTRD- and GeneHancer-annotated transcription factor–target gene network of untreated RA CD8+ Tem cells.
Table 16.
Top ten target genes in GTRD- and GeneHancer-annotated transcription factor–target gene network of untreated RA CD8+ Tem cells.
| Rank | Top Ten GTRD-Annotated Glycolysis Genes by In-Degree | Top Ten GeneHancer-Annotated Glycolysis Genes by In-Degree |
|---|
| Gene | DE | In-Degree in GTRD | In-Degree in GeneHancer | Common Edges * | Gene | DE | In-Degree in GTRD | In-Degree in GeneHancer | Common Edges * |
|---|
| 1 | PFKFB3 | DOWN | 103 | 79 | 53 | FBP2 | - | 11 | 103 | 5 |
| 2 | GAPDH | UP | 60 | 36 | 21 | PFKFB3 | DOWN | 103 | 79 | 53 |
| 3 | BPGM | - | 40 | 18 | 9 | DLD | DOWN | 39 | 36 | 20 |
| 4 | DLD | DOWN | 39 | 36 | 20 | GAPDH | UP | 60 | 36 | 21 |
| 5 | G6PD | - | 38 | 22 | 17 | ALDOB | - | 4 | 33 | 2 |
| 6 | SLC2A1 | - | 31 | 24 | 14 | G6PC2 | - | 3 | 24 | 1 |
| 7 | PDHA1 | - | 25 | 13 | 10 | SLC2A1 | - | 31 | 24 | 14 |
| 8 | PGAM1 | - | 18 | 11 | 7 | G6PC | - | 12 | 22 | 9 |
| 9 | AKR1A1 | - | 14 | 10 | 8 | G6PD | - | 38 | 22 | 17 |
| 10 | G6PC | - | 12 | 22 | 9 | HKDC1 | - | 6 | 19 | 4 |
Table 17.
Number of binding sites in GTRD and GeneHancer for PFKFB3, DLD and GAPDH in untreated RA CD8+ Tem cell differential co-expression networks.
Table 17.
Number of binding sites in GTRD and GeneHancer for PFKFB3, DLD and GAPDH in untreated RA CD8+ Tem cell differential co-expression networks.
| Gene | Number of Common Transcription Factors in GeneHancer and GTRD | Number of TFs with More Binding Sites in GeneHancer | Number of TFs with More Binding Sites in GTRD | Number of TFs with the Same Number of Binding Sites in GTRD and GeneHancer |
|---|
| PFKFB3 | 53 | 48 | 2 | 3 |
| GAPDH | 21 | 16 | 4 | 1 |
| DLD | 20 | 12 | 4 | 4 |
Table 18.
Differential expression of regulators of glycolysis in untreated RA CD8+ Tem cells. Means are calculated from DESeq2 normalized counts.
Table 18.
Differential expression of regulators of glycolysis in untreated RA CD8+ Tem cells. Means are calculated from DESeq2 normalized counts.
| Gene | adj. Pval | Log2FC | Linear FC | RA Mean | Control Mean |
|---|
| IRS2 | 0.072 | −1.268 | 0.415 | 40.38 | 97.49 |
| RUNX3 | 0.063 | −0.617 | 0.652 | 2925.192 | 4511.98 |
| ELOB | 0.003 | 1.766 | 3.401 | 2695.83 | 785.48 |
| EGLN3 | 0.073 | 1.616 | 3.064 | 27.06 | 8.55 |
| PFKFB3 | 0.00097 | −1.462 | 0.363 | 112.33 | 310.92 |
| PFKFB4 | 0.042 | 1.611 | 3.054 | 18.22 | 5.88 |
Table 19.
Top ten target genes in GTRD- and GeneHancer-annotated transcription factor–target gene network of untreated RA CD8+ Tem cells.
Table 19.
Top ten target genes in GTRD- and GeneHancer-annotated transcription factor–target gene network of untreated RA CD8+ Tem cells.
| Rank | Top Ten GTRD-Annotated Glycolysis Genes by In-Degree | Top Ten GeneHancer-Annotated Nodes by In-Degree |
|---|
| Gene | DE | In-Degree in GTRD | In-Degree in GeneHancer | Common Edges | Gene | DE | In-Degree in GTRD | In-Degree in GeneHancer | Common Edges |
|---|
| 1 | ENO1 | - | 189 | 135 | 88 | ENO1 | - | 189 | 135 | 88 |
| 2 | G6PC3 | - | 182 | 124 | 88 | G6PC3 | - | 182 | 124 | 88 |
| 3 | ALDOC | - | 173 | 59 | 28 | G6PC2 | - | 11 | 120 | 4 |
| 4 | SLC2A1 | - | 155 | 118 | 77 | AKR1A1 | - | 129 | 119 | 65 |
| 5 | ENO3 | UP | 150 | 106 | 76 | SLC2A1 | - | 155 | 118 | 77 |
| 6 | PDHA1 | UP | 147 | 72 | 43 | ADH7 | - | 60 | 109 | 17 |
| 7 | ACSS2 | - | 146 | 104 | 71 | HK2 | UP | 144 | 109 | 57 |
| 8 | HK2 | UP | 144 | 109 | 57 | ENO3 | UP | 150 | 106 | 76 |
| 9 | AKR1A1 | - | 129 | 119 | 65 | HK1 | - | 124 | 106 | 62 |
| 10 | HK1 | - | 124 | 106 | 62 | ACSS2 | - | 146 | 104 | 71 |
Table 20.
Expression levels of AKR1A1 and HK1 in untreated RA CD8+ Temra cells. Means are calculated from DESeq2 normalized counts.
Table 20.
Expression levels of AKR1A1 and HK1 in untreated RA CD8+ Temra cells. Means are calculated from DESeq2 normalized counts.
| Gene | adj. Pval | Log2FC | Linear FC | RA Mean | Control Mean |
|---|
| AKR1A1 | 0.05 | 0.56 | 1.48 | 92.08 | 60.7 |
| HK1 | 0.203279 | 0.533165 | 1.46 | 414.55 | 288.62 |
Table 21.
The differentially expressed genes in RA CD8+ Temra cells that are involved in the regulation of glycolysis. Means are calculated from DESeq2 normalized counts.
Table 21.
The differentially expressed genes in RA CD8+ Temra cells that are involved in the regulation of glycolysis. Means are calculated from DESeq2 normalized counts.
| Gene | adj. Pval | Log2FC | Linear FC | RA Mean | Control Mean |
|---|
| INSR | 0.090 | 1.92 | 3.77 | 12.191 | 2.947 |
| PPARG | 0.010 | 4.47 | 22.09 | 4.497 | 0.136 |
| MYC | 0.00045 | 1.657 | 3.14 | 153.618 | 45.369 |
| AKT1S1 | 0.086 | −0.78 | 0.58 | 29.087 | 50.460 |
| EGLN3 | 1.968 × 10−5 | 4.39 | 20.92 | 20.032 | 0.863 |
| HIF1AN | 0.00679 | 0.71 | 1.64 | 577.231 | 355.911 |
| EP300 | 0.0346 | 1.12 | 2.18 | 137.036 | 301.163 |