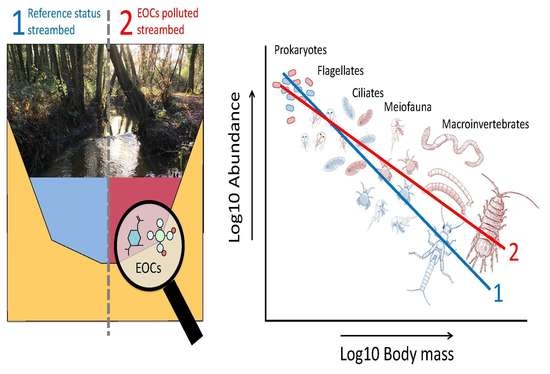
Mapping Micro-Pollutants and Their Impacts on the Size Structure of Streambed Communities
Abstract
:1. Introduction
2. Methods
2.1. Data Acquisition
2.2. Sampling and Processing of EOCs
2.3. Statistical Analysis
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vörösmarty, C.J.; McIntyre, P.B.; Gessner, M.O.; Dudgeon, D.; Prusevich, A.; Green, P.; Glidden, S.; Bunn, S.E.; Sullivan, C.A.; Liermann, C.R.; et al. Global threats to human water security and river biodiversity. Nature 2010, 467, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Schoups, G.; Van De Giesen, N. Organic pollution of rivers: Combined threats of urbanization, livestock farming and global climate change. Sci. Rep. 2017, 7, 43289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malaj, E.; Peter, C.; Grote, M.; Kühne, R.; Mondy, C.P.; Usseglio-Polatera, P. Organic chemicals jeopardize the health of freshwater ecosystems on the continental scale. Proc. Natl. Acad. Sci. USA 2014, 111, 9549–9554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarzenbach, R.P.; Escher, B.I.; Fenner, K.; Hofstetter, T.B.; Johnson, C.A.; Von Gunten, U.; Wehrli, B. The challenge of micropollutants in aquatic systems. Science 2006, 313, 1072–1077. [Google Scholar] [CrossRef] [PubMed]
- Reemtsma, T.; Weiss, S.; Mueller, J.; Petrovic, M.; González, S.; Barcelo, D.; Ventura, F.; Knepper, T.P. Polar pollutants entry into the water cycle by municipal wastewater: A European perspective. Environ. Sci. Technol. 2006, 40, 5451–5458. [Google Scholar] [CrossRef] [PubMed]
- Pal, A.; Gin, K.Y.H.; Lin, A.Y.C.; Reinhard, M. Impacts of emerging organic contaminants on freshwater resources: Review of recent occurrences, sources, fate and effects. Sci. Total Environ. 2010, 408, 6062–6069. [Google Scholar] [CrossRef] [PubMed]
- Kidd, K.A.; Blanchfield, P.J.; Mills, K.H.; Palace, V.P.; Evans, R.E.; Lazorchak, J.M.; Flick, R.W. Collapse of a fish population after exposure to a synthetic estrogen. Proc. Natl. Acad. Sci. USA 2007, 104, 8897–8901. [Google Scholar] [CrossRef] [Green Version]
- Galus, M.; Rangarajan, S.; Lai, A.; Shaya, L.; Balshine, S.; Wilson, J.Y. Effects of chronic, parental pharmaceutical exposure on zebrafish (Danio rerio) offspring. Aquat. Toxicol. 2014, 151, 124–134. [Google Scholar] [CrossRef]
- Stamm, C.; Räsänen, K.; Burdon, F.J.; Altermatt, F.; Jokela, J.; Joss, A.; Ackermann, M.; Eggen, R.I. Unravelling the impacts of micropollutants in aquatic ecosystems: Interdisciplinary studies at the interface of large-scale ecology. Adv. Ecol. Res. 2016, 55, 183–223. [Google Scholar]
- Kubec, J.; Hossain, S.; Grabicová, K.; Randák, T.; Kouba, A.; Grabic, R.; Roje, S.; Buřič, M. Oxazepam alters the behavior of crayfish at diluted concentrations, venlafaxine does not. Water 2019, 11, 196. [Google Scholar] [CrossRef] [Green Version]
- Lewandowski, J.; Arnon, S.; Banks, E.; Batelaan, O.; Betterle, A.; Broecker, T.; Coll, C.; Drummond, J.D.; Garcia, J.G.; Galloway, J.; et al. Is the Hyporheic Zone Relevant beyond the Scientific Community? Water 2019, 11, 2230. [Google Scholar] [CrossRef] [Green Version]
- Posselt, M.; Jaeger, A.; Schaper, J.L.; Radke, M.; Benskin, J.P. Determination of polar organic micropollutants in surface and pore water by high-resolution sampling-direct injection-ultra high performance liquid chromatography-tandem mass spectrometry. Environ. Sci. Process. Impacts 2018, 20, 1716–1727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richmond, E.K.; Grace, M.R.; Kelly, J.J.; Reisinger, A.J.; Rosi, E.J.; Walters, D.M. Pharmaceuticals and personal care products (PPCPs) are ecological disrupting compounds (EcoDC). Elem. Sci. Anth. 2017, 5, 66. [Google Scholar] [CrossRef] [Green Version]
- Findlay, S. Importance of surface-subsurface exchange in stream ecosystems: The hyporheic zone. Limnol. Oceanogr. 1995, 40, 159–164. [Google Scholar] [CrossRef]
- Hester, E.T.; Young, K.I.; Widdowson, M.A. Mixing of surface and groundwater induced by riverbed dunes: Implications for hyporheic zone definitions and pollutant reactions. Water Resour. Res. 2013, 49, 5221–5237. [Google Scholar] [CrossRef]
- Subirats, J.; Triadó-Margarit, X.; Mandaric, L.; Acuña, V.; Balcázar, J.L.; Sabater, S.; Borrego, C.M. Wastewater pollution differently affects the antibiotic resistance gene pool and biofilm bacterial communities across streambed compartments. Mol. Ecol. 2017, 26, 5567–5581. [Google Scholar] [CrossRef] [Green Version]
- Subirats, J.; Timoner, X.; Sànchez-Melsió, A.; Balcázar, J.L.; Acuña, V.; Sabater, S.; Borrego, C.M. Emerging contaminants and nutrients synergistically affect the spread of class 1 integron-integrase (intI1) and sul1 genes within stable streambed bacterial communities. Water Res. 2018, 138, 77–85. [Google Scholar] [CrossRef]
- Láng, J.; Kőhidai, L. Effects of the aquatic contaminant human pharmaceuticals and their mixtures on the proliferation and migratory responses of the bioindicator freshwater ciliate Tetrahymena. Chemosphere 2012, 89, 592–601. [Google Scholar] [CrossRef]
- Althakafy, J.T.; Kulsing, C.; Grace, M.R.; Marriott, P.J. Determination of selected emerging contaminants in freshwater invertebrates using a universal extraction technique and liquid chromatography accurate mass spectrometry. J. Sep. Sci. 2018, 41, 3706–3715. [Google Scholar] [CrossRef]
- Miller, T.H.; Ng, K.T.; Bury, S.T.; Bury, S.E.; Bury, N.R.; Barron, L.P. Biomonitoring of pesticides, pharmaceuticals and illicit drugs in a freshwater invertebrate to estimate toxic or effect pressure. Environ. Int. 2019, 129, 595–606. [Google Scholar] [CrossRef]
- Peralta-Maraver, I.; Perkins, D.M.; Thompson, M.S.; Fussmann, K.; Reiss, J.; Robertson, A.L. Comparing biotic drivers of litter breakdown across stream compartments. J. An. Ecol. 2019, 88, 1146–1157. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.H.; Gillooly, J.F.; Allen, A.P.; Savage, V.M.; West, G.B. Toward a metabolic theory of ecology. Ecology 2004, 85, 1771–1789. [Google Scholar] [CrossRef]
- White, E.P.; Ernest, S.M.; Kerkhoff, A.J.; Enquist, B.J. Relationships between body size and abundance in ecology. Trends Ecol. Evol. 2007, 22, 323–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trebilco, R.; Baum, J.K.; Salomon, A.K.; Dulvy, N.K. Ecosystem ecology: Size-based constraints on the pyramids of life. Trends Ecol. Evol. 2013, 28, 423–431. [Google Scholar] [CrossRef]
- Kerr, S.R.; Dickie, L.M. The Biomass Spectrum: A PredatorPrey Theory of Aquatic Production; Columbia University Press: Chichester, NY, USA, 2001. [Google Scholar]
- Perkins, D.M.; Durance, I.; Edwards, F.K.; Grey, J.; Hildrew, A.G.; Jackson, M.; Jones, J.I.; Lauridsen, R.B.; Layer-Dobra, K.; Thompson, M.S.; et al. Bending the rules: Exploitation of allochthonous resources by a top-predator modifies size-abundance scaling in stream food webs. Ecol. Lett. 2018, 21, 1771–1780. [Google Scholar] [CrossRef] [Green Version]
- Peralta-Maraver, I.; Robertson, A.L.; Perkins, D.M. Depth and vertical hydrodynamics constrain the size structure of a lowland streambed community. Biol. Lett. 2019, 15, 20190317. [Google Scholar] [CrossRef] [Green Version]
- Petchey, O.L.; Morin, P.J.; Hulot, F.D. Contributions of aquatic model systems to our understanding of biodiversity and ecosystem functioning. In Biodiversity and Ecosystem Functioning—Synthesis and Perspectives; Loreau, M., Naeem, S., Inchausti, P., Eds.; Oxford University Press: Oxford, UK, 2002; pp. 127–138. [Google Scholar]
- Petchey, O.L.; Belgrano, A. Body-size distributions and size-spectra: Universal indicators of ecological status? Biol. Lett. 2010, 6, 434–437. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Sobek, A.; Radke, M. Fate of pharmaceuticals and their transformation products in four small European rivers receiving treated wastewater. Environ. Sci. Technol. 2016, 50, 5614–5621. [Google Scholar] [CrossRef] [Green Version]
- Schaper, J.L.; Posselt, M.; Bouchez, C.; Jaeger, A.; Nuetzmann, G.; Putschew, A.; Singer, G.; Lewandowski, J. Fate of Trace Organic Compounds in the Hyporheic Zone: Influence of Retardation, the Benthic Biolayer, and Organic Carbon. Environ. Sci. Technol. 2019, 53, 4224–4234. [Google Scholar] [CrossRef]
- Mechelke, J.; Vermeirssen, E.L.; Hollender, J. Passive sampling of organic contaminants across the water-sediment interface of an urban stream. Water Res. 2019, 165, 114966. [Google Scholar] [CrossRef] [Green Version]
- Bray, J.R.; Curtis, J.T. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Oksanen, J. Multivariate Analysis of Ecological Communities in R: Vegan Tutorial. 2015. Available online: http://cc.oulu.fi/~jarioksa/opetus/metodi/veg-antutor.pdf (accessed on 19 November 2019).
- Peralta-Maraver, I.; Robertson, A.L.; Rezende, E.L.; Lemes da Silva, A.L.; Tonetta, D.; Lopes, M.; Schmitt, R.; Leite, N.K.; Nuñer, A.; Petrucio, M.M. Winter is coming: Food web structure and seasonality in a subtropical freshwater coastal lake. Ecol. Evol. 2017, 7, 4534–4542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edwards, A.M.; Robinson, J.P.; Plank, M.J.; Baum, J.K.; Blanchard, J.L. Testing and recommending methods for fitting size spectra to data. Methods Ecol. Evol. 2017, 8, 57–67. [Google Scholar] [CrossRef]
- Mulder, C.; Elser, J.J. Soil acidity, ecological stoichiometry and allometric scaling in grassland food webs. Glob. Chang. Biol. 2009, 15, 2730–2738. [Google Scholar] [CrossRef] [Green Version]
- Layer, K.; Riede, J.O.; Hildrew, A.G.; Woodward, G. Food web structure and stability in 20 streams across a wide ph gradient. Adv. Ecol. Res. 2010, 42, 265–299. [Google Scholar]
- Zuur, A.F.; Ieno, E.N.; Walker, N.J.; Savaliev, A.A.; Smith, G.M. Mixed Effects Models and Extensions in Ecology with R; Springer: New York, NY, USA, 2009; p. 57. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community Ecology Package. R Package Version 2.0–10. 2013. Available online: http://CRAN.R–project.org/ package=vegan (accessed on 7 November 2019).
- Dossena, M.; Yvon-Durocher, G.; Grey, J.; Montoya, J.M.; Perkins, D.M.; Trimmer, M.; Woodward, G. Warming alters community size structure and ecosystem functioning. Proc. Biol. Sci. Replaces Proc. R. Soc. 2012, 279, 3011–3019. [Google Scholar] [CrossRef] [Green Version]
- O’Gorman, E.J.; Pichler, D.E.; Adams, G.; Benstead, J.P.; Cohen, H.; Craig, N.; Cross, W.F.; Demars, B.O.; Friberg, N.; Gislason, G.M.; et al. Impacts of warming on the structure and functioning of aquatic communities: Individual-to ecosystem-level responses. Adv. Ecol. Res. 2012, 47, 81–176. [Google Scholar]
- Hawkes, H.A. Origin and development of the biological monitoring working party score system. Water Res. 1998, 32, 964–968. [Google Scholar] [CrossRef]
- Zenker, A.; Cicero, M.R.; Prestinaci, F.; Bottoni, P.; Carere, M. Bioaccumulation and biomagnification potential of pharmaceuticals with a focus to the aquatic environment. J. Environ. Manag. 2014, 133, 378–387. [Google Scholar] [CrossRef]
- Ruhí, A.; Acuña, V.; Barceló, D.; Huerta, B.; Mor, J.R.; Rodríguez-Mozaz, S.; Sabater, S. Bioaccumulation and trophic magnification of pharmaceuticals and endocrine disruptors in a Mediterranean river food web. Sci. Total Environ. 2016, 540, 250–259. [Google Scholar] [CrossRef]
- Richmond, E.K.; Rosi, E.J.; Walters, D.M.; Fick, J.; Hamilton, S.K.; Brodin, T.; Sundelin, A.; Grace, M.R. A diverse suite of pharmaceuticals contaminates stream and riparian food webs. Nat. Commun. 2019, 9, 4491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, J.N.; Paxéus, N.; Förlin, L.; Larsson, D.J. Variations in bioconcentration of human pharmaceuticals from sewage effluents into fish blood plasma. Environ. Toxicol. Pharmacol. 2007, 24, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Fick, J.; Lindberg, R.H.; Tysklind, M.; Larsson, D.J. Predicted critical environmental concentrations for 500 pharmaceuticals. Regul. Toxicol. Pharmacol. 2010, 58, 516–523. [Google Scholar] [CrossRef] [PubMed]
- Buerge, I.J.; Buser, H.-R.; Kahle, M.; Müller, M.D.; Poiger, T. Ubiquitous occurrence of the artificial sweetener acesulfame in the aquatic environment: An ideal chemical marker of domestic wastewater in groundwater. Environ. Sci. Technol. 2009, 43, 4381–4385. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, A.; Posselt, M.; Betterle, A.; Schaper, J.; Mechelke, J.; Coll, C.; Lewandowski, J. Spatial and Temporal Variability in Attenuation of Polar Organic Micropollutants in an Urban Lowland Stream. Environ. Sci. Technol. 2019, 53, 2383–2395. [Google Scholar] [CrossRef] [Green Version]
- Jaeger, A.; Coll, C.; Posselt, M.; Mechelke, J.; Rutere, C.; Betterle, A.; Raza, M.; Mehrtens, A.; Meinikmann, K.; Portmann, A.; et al. Using recirculating flumes and a response surface model to investigate the role of hyporheic exchange and bacterial diversity on micropollutant. Environ. Sci. Process. Impacts 2019, 21. [Google Scholar] [CrossRef] [Green Version]


| River | Lat | Lon | Acesu | Aceta | Sita | O.Des | 11.D.C. | Napro | Guan | Metf | Venl | M.Acid | Carba | Oxa | Prop | Sota | 4.H.D. | Trama | Diclo | 1.H.B. | Ibu | 11.D.C. | C.Acid2 | Furo | Meto | Gem | Tot EOCs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beverly Brooks | 51.44 | 0.25 | 13.8 | 0.1 | 0.4 | 1.1 | 44.8 | 1.5 | 0.6 | 0.6 | 0.2 | 0.6 | 0.1 | 0.2 | 2.9 | 0.6 | 14 | ||||||||||
| Loddon | 51.42 | 1.72 | 0.8 | 0.1 | 0.4 | 0.8 | 9.7 | 0.2 | 0.2 | 0.1 | 0.2 | 0.7 | 0.1 | 0.4 | 0.1 | 13 | |||||||||||
| Wey | 51.19 | 0.68 | 0.8 | 0.5 | 0.1 | 0.1 | 0.7 | 5.2 | 0.4 | 0.1 | 0.8 | 0.3 | 0.2 | 0.6 | 12 | ||||||||||||
| Waveney | 52.42 | 1.36 | 0.5 | 0.1 | 0.6 | 0.2 | 1.7 | 0.4 | 0.2 | 0.3 | 0.1 | 9 | |||||||||||||||
| Wensum | 52.42 | 1.36 | 0.2 | 0.4 | 0.3 | 0.4 | 0.4 | 0.8 | 0.2 | 0.3 | 8 | ||||||||||||||||
| Deadwater | 51.17 | 0.85 | 1.9 | 0.2 | 0.7 | 0.1 | 0.7 | 0.1 | 6 | ||||||||||||||||||
| Stiffkey | 52.92 | 0.89 | 0.3 | 0.2 | 0.2 | 0.2 | 0.1 | 5 | |||||||||||||||||||
| Tat | 52.82 | 0.75 | 0.5 | 0.9 | 0.2 | 0.9 | 4 | ||||||||||||||||||||
| River Leith | 54.61 | −2.62 | 0.5 | 0.4 | 0.4 | 3 | |||||||||||||||||||||
| Nadder | 51.12 | 0.90 | 1.7 | 0.4 | 0.1 | 3 | |||||||||||||||||||||
| Test | 51.14 | 1.47 | 1.0 | 0.2 | 0.4 | 3 | |||||||||||||||||||||
| Glaven | 52.93 | 1.63 | 0.2 | 0.7 | 0.4 | 3 | |||||||||||||||||||||
| Lamports | 51.15 | 1.72 | 0.1 | 0.3 | 0.1 | 3 | |||||||||||||||||||||
| Lyde | 51.29 | 1.72 | 0.2 | 0.8 | 0.2 | 3 | |||||||||||||||||||||
| GI1 | 52.14 | −3.84 | 0.4 | 0.1 | 2 | ||||||||||||||||||||||
| Howe Beck | 54.68 | −2.59 | 0.1 | 0.7 | 2 | ||||||||||||||||||||||
| Bure | 52.82 | 1.21 | 0.1 | 0.1 | 2 | ||||||||||||||||||||||
| River Crowdundle | 51.15 | 1.72 | 0.1 | 0.1 | 2 | ||||||||||||||||||||||
| Kennet | 51.42 | 1.72 | 0.2 | 0.7 | 2 | ||||||||||||||||||||||
| River Lyvennet | 54.68 | −2.61 | 0.1 | 0.5 | 2 | ||||||||||||||||||||||
| LI7 | 52.13 | −3.75 | 0 | ||||||||||||||||||||||||
| LI8 | 52.16 | −3.75 | 0 | ||||||||||||||||||||||||
| LI3 | 52.14 | −3.73 | 0 | ||||||||||||||||||||||||
| Old Lodge | 54.65 | −2.64 | 0 | ||||||||||||||||||||||||
| Lone Oak | 51.77 | 0.13 | 0 | ||||||||||||||||||||||||
| LI6 | 51.44 | 0.25 | 0 | ||||||||||||||||||||||||
| Broadstone Stream | 51.89 | 0.57 | 0 | ||||||||||||||||||||||||
| Oakhanger | 51.45 | 0.79 | 0 | ||||||||||||||||||||||||
| Anton | 51.15 | 1.46 | 0 | ||||||||||||||||||||||||
| Morland Beck | 51.23 | 1.72 | 0 |
| Response | Predictors | N | AIC | ΔAIC | LogLik | wi |
|---|---|---|---|---|---|---|
| Log 10 (N) | log10(M) × EOCs + pH + Temp + Lon + Lat + Alt + Nit + Phos | 12 | 403.82 | 9.23 | 0.01 | 0.00 |
| log10(M) × EOCs + pH + Temp + Lon + Lat + Alt + Nit | 11 | 402.30 | 7.70 | 0.02 | 0.01 | |
| log10(M) × EOCs + pH + Temp + Lon + Lat + Alt | 10 | 402.35 | 7.75 | 0.02 | 0.01 | |
| log10(M) × EOCs + pH + Temp + Lon + Lat | 9 | 401.17 | 6.57 | 0.04 | 0.02 | |
| log10(M) × EOCs + pH + Temp + Lon | 8 | 399.18 | 4.59 | 0.10 | 0.05 | |
| log10(M) × EOCs + pH + Temp | 7 | 397.32 | 2.72 | 0.26 | 0.12 | |
| log10(M) × EOCs + pH | 6 | 396.15 | 1.55 | 0.46 | 0.22 | |
| log10(M) × EOCs | 5 | 394.60 | 0.00 | 1.00 | 0.47 | |
| log10(M) + EOCs | 4 | 397.88 | 3.28 | 0.19 | 0.09 | |
| log10(M) | 3 | 401.22 | 6.62 | 0.04 | 0.02 |
| Fixed Equation Terms | Coef | SE | t Value | p Value | Sig |
|---|---|---|---|---|---|
| Intercept | 2.95 | 0.12 | 25.20 | > 0.001 | *** |
| Log10 body mass | −0.37 | 0.03 | −13.87 | > 0.001 | *** |
| Presence/absence of EOCs | −0.77 | 0.24 | −3.22 | 0.001 | *** |
| Log10 body mass * EOC pollution | −0.12 | 0.05 | 0.05 | 0.02 | * |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peralta-Maraver, I.; Posselt, M.; Perkins, D.M.; Robertson, A.L. Mapping Micro-Pollutants and Their Impacts on the Size Structure of Streambed Communities. Water 2019, 11, 2610. https://doi.org/10.3390/w11122610
Peralta-Maraver I, Posselt M, Perkins DM, Robertson AL. Mapping Micro-Pollutants and Their Impacts on the Size Structure of Streambed Communities. Water. 2019; 11(12):2610. https://doi.org/10.3390/w11122610
Chicago/Turabian StylePeralta-Maraver, Ignacio, Malte Posselt, Daniel M. Perkins, and Anne L. Robertson. 2019. "Mapping Micro-Pollutants and Their Impacts on the Size Structure of Streambed Communities" Water 11, no. 12: 2610. https://doi.org/10.3390/w11122610
APA StylePeralta-Maraver, I., Posselt, M., Perkins, D. M., & Robertson, A. L. (2019). Mapping Micro-Pollutants and Their Impacts on the Size Structure of Streambed Communities. Water, 11(12), 2610. https://doi.org/10.3390/w11122610