Automated Targeted Sampling of Waterborne Pathogens and Microbial Source Tracking Markers Using Near-Real Time Monitoring of Microbiological Water Quality
Abstract
:1. Introduction
2. Material and Methods
2.1. Sampling Sites
2.2. Online Monitoring of β-D-Glucuronidase (GLUC) Activity
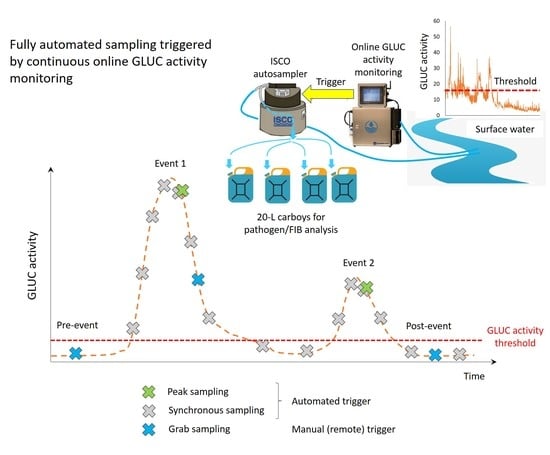
2.3. Triggering ISCO Autosampler by Online In Situ GLUC Activity Measurements
2.3.1. Communication between ISCO Autosampler and GLUC Activity Online Monitoring Instrument
- UnixTrigger: Unix timestamp when sampling starts;
- UnixFinished: Unix timestamp when sampling is completed;
- UID (unique identifier): UID of the measurement that triggered the ISCO (if trigger was not manual);
- Status message
- ○
- SUCCESS (0): Sampling successful;
- ○
- PROGINACTIVE (1): Program on ISCO not started or not started/configured correctly;
- ○
- RUNTIMEERROR (2): ISCO reports runtime errors during sampling;
- ○
- PUMPERROR (3): ISCO reports pump faults;
- ○
- NOUSBDEVICE (4): FTDI chip not found—ISCO module not connected;
- ○
- PROGNOTFOUND (5): Program for ISCO control not found/not installed.
2.3.2. Triggering and Sampling Modes
- Manual trigger, either on-site or through remote control.
- Automated trigger, synchronous with the next autonomous measurement of GLUC activity.
- Automated trigger, immediately when a GLUC activity measurement result is provided (15 min after sampling for GLUC activity measurement) and a predefined trigger condition is fulfilled. For sampling at the peak of a GLUC activity pollutograph, the predefined conditions were set as follows:
- ○
- Below GLUC activity (GA) threshold: no trigger;
- ○
- Above GA threshold:
- ▪
- If GAn ≥ GAn − 1: no trigger;
- ▪
- If GAn < GAn − 1: immediate trigger of ISCO autosampler upon GAn result acquisition (15 min after sampling for GLUC activity measurement);
- ○
- Above GA threshold and after the peak GA (GAmax):
- ▪
- If GAn exceeds GAmax: the same algorithms described above do apply.
2.3.3. Field Validation of the Triggering Modes
2.4. Microbiological Analyses
2.4.1. Culture-Based Enumeration of E. coli
2.4.2. Enumeration of Protozoan Parasites
2.4.3. Bacteroides Quantification
2.5. Hydrometeorological Measurements and Online Physico-Chemistry
3. Results and Discussion
3.1. Synchronous Sampling
3.2. Peak Sampling
3.3. Event-Based Sampling Triggered by Online Monitoring of Microbiological Water Quality
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Quantitative Microbial Risk Assessment: Application for Water Safety Management; WHO Press: Geneva, Switzerland, 2016. [Google Scholar]
- Besmer, M.D.E.; Epting, J.; Page, R.M.; Sigrist, J.A.; Huggenberger, P.; Hammes, F. Online flow cytometry reveals microbial dynamics influenced by concurrent natural and operational events in groundwater used for drinking water treatment. Sci. Rep. 2016, 6, 38462. [Google Scholar] [CrossRef]
- Boehm, A.B. Enterococci concentrations in diverse coastal environments exhibit extreme variability. Environ. Sci. Technol. 2007, 41, 8227–8232. [Google Scholar] [CrossRef] [Green Version]
- Burnet, J.-B.; Penny, C.; Ogorzaly, L.; Cauchie, H.-M. Spatial and temporal distribution of Cryptosporidium and Giardia in a drinking water resource: Implications for monitoring and risk assessment. Sci. Total. Environ. 2014, 472, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Dorner, S.; Anderson, W.; Gaulin, T.; Candon, H.; Slawson, R.; Payment, P.; Huck, P. Pathogen and indicator variability in a heavily impacted watershed. J. Water Health 2007, 5, 241–257. [Google Scholar] [CrossRef]
- Partyka, M.L.; Bond, R.F.; Chase, J.A.; Atwill, E.R. Spatial and temporal variability of bacterial indicators and pathogens in six California reservoirs during extreme drought. Water Res. 2018, 129, 436–446. [Google Scholar] [CrossRef] [PubMed]
- Madrid, Y.; Zayas, Z.P. Water sampling: Traditional methods and new approaches in water sampling strategy. TrAC Trends Anal. Chem. 2007, 26, 293–299. [Google Scholar] [CrossRef]
- Hrudey, S.E.; Hrudey, E.J. Common themes contributing to recent drinking water disease outbreaks in affluent nations. Water Supply 2019, 19, 1767–1777. [Google Scholar] [CrossRef] [Green Version]
- Moreira, N.A.; Bondelind, M. Safe drinking water and waterborne outbreaks. J. Water Health 2016, 15, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Cann, K.F.; Thomas, D.R.; Salmon, R.L.; Wyn-Jones, A.P.; Kay, D. Extreme water-related weather events and waterborne disease. Epidemiol. Infect. 2012, 141, 671–686. [Google Scholar] [CrossRef]
- Soller, J.A.; Schoen, M.; Steele, J.A.; Griffith, J.F.; Schiff, K.C. Incidence of gastrointestinal illness following wet weather recreational exposures: Harmonization of quantitative microbial risk assessment with an epidemiologic investigation of surfers. Water Res. 2017, 121, 280–289. [Google Scholar] [CrossRef]
- USEPA. Occurrence and Exposure Assessment for the Final Long Term Enhanced Surface Water Treatment Rule; EPA 815-R-06-002; US Environmental Protection Agency: Washington, DC, USA, 2005.
- Gouvernement du Québec. Règlement sur le Prélèvement des Eaux et Leur Protection (Québec, Canada); Gouvernement du Québec: Quebec City, QC, Canada, 2018. [Google Scholar]
- EC. European Council Directive 2006/7/EC of 15 February 2006 Concerning the Management of Bathing Water Quality and Repealing Directive 76/160/EEC; Official Journal of the European Union: Strasbourg, France, 2006. [Google Scholar]
- USEPA. Recreational Water Quality Criteria; EPA 820-F-12-058 U.S.; Environmental Protection Agency: Washington, DC, USA, 2012; pp. 1–69.
- Besmer, M.D.; Hammes, F.; Sigrist, J.A.; Ort, C. Evaluating monitoring strategies to detect precipitation-induced microbial contamination events in karstic springs used for drinking water. Front. Microbiol. 2017, 8, 2229. [Google Scholar] [CrossRef]
- Roig, B.; Delpla, I.; Baurès, E.; Jung, A.; Thomas, O. Analytical issues in monitoring drinking-water contamination related to short-term, heavy rainfall events. TrAC Trends Anal. Chem. 2011, 30, 1243–1251. [Google Scholar] [CrossRef]
- Lewis, J. Turbidity-controlled suspended sediment sampling for runoff-event load estimation. Water Resour. Res. 1996, 32, 2299–2310. [Google Scholar] [CrossRef]
- Åström, J.; Pettersson, T.; Reischer, G.H.; Hermansson, M. Short-term microbial release during rain events from on-site sewers and cattle in a surface water source. J. Water Health 2013, 11, 430–442. [Google Scholar] [CrossRef] [PubMed]
- Signor, R.S.; Roser, D.; Ashbolt, N.; Ball, J. Quantifying the impact of runoff events on microbiological contaminant concentrations entering surface drinking source waters. J. Water Health 2005, 3, 453–468. [Google Scholar] [CrossRef] [Green Version]
- Stadler, H.; Skritek, P.; Sommer, R.; Mach, R.; Zerobin, W.; Farnleitner, A. Microbiological monitoring and automated event sampling at karst springs using LEO-satellites. Water Sci. Technol. 2008, 58, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Burnet, J.-B.; Sylvestre, É.; Jalbert, J.; Imbeault, S.; Servais, P.; Prévost, M.; Dorner, S. Tracking the contribution of multiple raw and treated wastewater discharges at an urban drinking water supply using near real-time monitoring of β-d-glucuronidase activity. Water Res. 2019, 164, 114869. [Google Scholar] [CrossRef] [PubMed]
- Sylvestre, É.; Burnet, J.; Dorner, S.; Smeetsc, P.; Medema, G.; Villion, M.; Hachad, M.; Prévost, M. Impact of hydrometeorological events for the selection of parametric models for protozoan pathogens in drinking-water sources. Risk Anal. 2020, 13612. [Google Scholar] [CrossRef]
- Burnet, J.B.; Dinh, Q.T.; Imbeault, S.; Servais, P.; Dorner, S.; Prévost, M. Autonomous online measurement of b-D-glucuronidase activity in surface water: Is it suitable for rapid E. coli monitoring? Water Res. 2019, 152, 241–250. [Google Scholar] [CrossRef]
- Buysschaert, B.; Vermijs, L.; Naka, A.; Boon, N.; De Gusseme, B. Online flow cytometric monitoring of microbial water quality in a full-scale water treatment plant. npj Clean Water 2018, 1, 16. [Google Scholar] [CrossRef] [Green Version]
- Demeter, K.; Burnet, J.-B.; Stadler, P.; Kirschner, A.; Zessner, M.; Farnleitner, A.H. Automated online monitoring of fecal pollution in water by enzymatic methods. Curr. Opin. Environ. Sci. Health 2020, 16, 82–91. [Google Scholar] [CrossRef]
- Sorensen, J.; Vivanco, A.; Ascott, M.; Gooddy, D.; Lapworth, D.; Read, D.; Rushworth, C.; Bucknall, J.; Herbert, K.; Karapanos, I.; et al. Online fluorescence spectroscopy for the real-time evaluation of the microbial quality of drinking water. Water Res. 2018, 137, 301–309. [Google Scholar] [CrossRef]
- Stadler, P.; Bloschl, G.; Vogl, W.; Koschelnik, J.; Epp, M.; Lackner, M.; Oismüller, M.; Kumpan, M.; Nemeth, L.; Strauss, P.; et al. Real-time monitoring of beta-d-glucuronidase activity in sediment laden streams: A comparison of prototypes. Water Res. 2016, 101, 252–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ender, A.; Goeppert, N.; Grimmeisen, F.; Goldscheider, N. Evaluation of β-d-glucuronidase and particle-size distribution for microbiological water quality monitoring in Northern Vietnam. Sci. Total. Environ. 2017, 580, 996–1006. [Google Scholar] [CrossRef]
- Rice, E.W.; Allen, M.J. Efficacy of beta-glucuronidase assay for identification of Escherichia coli by the defined-substrate technology. Appl. Environ. Microbiol. 1990, 56, 1203–1205. [Google Scholar] [CrossRef] [Green Version]
- Servais, P.; Prats, J.; Passerat, J.; Garcia-Armisen, T. Abundance of culturable versus viable Escherichia coli in freshwater. Can. J. Microbiol. 2009, 55, 905–909. [Google Scholar] [CrossRef]
- Cazals, M.; Stott, R.; Fleury, C.; Proulx, F.; Prévost, M.; Servais, P.; Dorner, S.; Burnet, J.-B. Near real-time notification of water quality impairments in recreational freshwaters using rapid online detection of β-D-glucuronidase activity as a surrogate for Escherichia coli monitoring. Sci. Total. Environ. 2020, 720, 137303. [Google Scholar] [CrossRef] [PubMed]
- Koschelnik, J.; Vogl, W.; Epp, M.; Lackner, M. Rapid analysis of β-D-glucuronidase activity in water using fully automated technology. In Water Resources Management VIII; WIT Press: Southamption, UK, 2015; pp. 471–481. [Google Scholar]
- USEPA. Method 1604: Total Coliforms and Escherichia coli in Water by Membrane Filtration Using a Simultaneous Detection Technique (MI Medium); USEPA: Washington, DC, USA, 2002. [Google Scholar]
- Dick, L.K.; Field, K.G. Rapid estimation of numbers of fecal Bacteroidetes by use of a quantitative PCR assay for 16s rRNA genes. Appl. Environ. Microbiol. 2004, 70, 5695–5697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ontario Ministry of the Environment Conservation and Parks. Identification of Fecal Waste Pollution Sources by Real-Time Polymerase Chain Reaction (PCR) Using Bacteroidales 16S rRNA Genetic Markers; Ontario Ministry of the Environment Conservation and Parks: Toronto, ON, Canada, 2016. [Google Scholar]
- Lee, D.-Y.; Weir, S.C.; Lee, H.; Trevors, J.T. Quantitative identification of fecal water pollution sources by TaqMan real-time PCR assays using Bacteroidales 16S rRNA genetic markers. Appl. Microbiol. Biotechnol. 2010, 88, 1373–1383. [Google Scholar] [CrossRef]
- Lee, D.-Y.; Lee, H.; Trevors, J.T.; Weir, S.C.; Thomas, J.L.; Habash, M. Characterization of sources and loadings of fecal pollutants using microbial source tracking assays in urban and rural areas of the Grand River Watershed, Southwestern Ontario. Water Res. 2014, 53, 123–131. [Google Scholar] [CrossRef]
- Environment Canada. Available online: https://eau.ec.gc.ca/mainmenu/historical_data_index_f.html (accessed on 27 July 2021).
- Sylvestre, É.; Burnet, J.-B.; Smeets, P.; Medema, G.; Prévost, M.; Dorner, S. Can routine monitoring of E. coli fully account for peak event concentrations at drinking water intakes in agricultural and urban rivers? Water Res. 2020, 170, 115369. [Google Scholar] [CrossRef] [PubMed]
- Huey, G.M.; Meyer, M.L. Turbidity as an indicator of water quality in diverse watersheds of the Upper Pecos River basin. Water 2010, 2, 273–284. [Google Scholar] [CrossRef]
- Farnleitner, A.; Ryzinska-Paier, G.; Reischer, G.; Burtscher, M.; Knetsch, S.; Kirschner, A.; Dirnböck, T.; Kuschnig, G.; Mach, R.; Sommer, R. Escherichia coli and enterococci are sensitive and reliable indicators for human, livestock and wildlife faecal pollution in alpine mountainous water resources. J. Appl. Microbiol. 2010, 109, 1599–1608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atherholt, T.B.; Lechevallier, M.W.; Norton, W.D.; Rosen, J.S. Effect of rainfall on Giardia and crypto. J. Am. Water Work. Assoc. 1998, 90, 66–80. [Google Scholar] [CrossRef]
- Dechesne, M.; Soyeux, E. Assessment of source water pathogen contamination. J. Water Health. 2007, 5, 39–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kistemann, T.; Claßen, T.; Koch, C.; Dangendorf, F.; Fischeder, R.; Gebel, J.; Vacata, V.; Exner, M. Microbial load of drinking water reservoir tributaries during extreme rainfall and runoff. Appl. Environ. Microbiol. 2002, 68, 2188–2197. [Google Scholar] [CrossRef] [Green Version]
- Janke, R.; Murray, R.; Uber, J.; Taxon, T. Comparison of physical sampling and real-time monitoring strategies for designing a contamination warning system in a drinking water distribution system. J. Water Resour. Plan. Manag. 2006, 132, 310–313. [Google Scholar] [CrossRef]
- Madoux-Humery, A.S.A.; Dorner, S.S.; Sauvé, S.; Aboulfadl, K.; Galarneau, M.; Servais, P.; Prévost, M. The effects of combined sewer overflow events on riverine sources of drinking water. Water Res. 2016, 92, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Passerat, J.; Ouattara, N.K.; Mouchel, J.-M.; Rocher, V.; Servais, P. Impact of an intense combined sewer overflow event on the microbiological water quality of the Seine River. Water Res. 2011, 45, 893–903. [Google Scholar] [CrossRef]
- McCarthy, D.T.; Zhang, K.; Westerlund, C.; Viklander, M.; Bertrand-Krajewski, J.-L.; Fletcher, T.D.; Deletic, A. Assessment of sampling strategies for estimation of site mean concentrations of stormwater pollutants. Water Res. 2018, 129, 297–304. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Burnet, J.-B.; Habash, M.; Hachad, M.; Khanafer, Z.; Prévost, M.; Servais, P.; Sylvestre, E.; Dorner, S. Automated Targeted Sampling of Waterborne Pathogens and Microbial Source Tracking Markers Using Near-Real Time Monitoring of Microbiological Water Quality. Water 2021, 13, 2069. https://doi.org/10.3390/w13152069
Burnet J-B, Habash M, Hachad M, Khanafer Z, Prévost M, Servais P, Sylvestre E, Dorner S. Automated Targeted Sampling of Waterborne Pathogens and Microbial Source Tracking Markers Using Near-Real Time Monitoring of Microbiological Water Quality. Water. 2021; 13(15):2069. https://doi.org/10.3390/w13152069
Chicago/Turabian StyleBurnet, Jean-Baptiste, Marc Habash, Mounia Hachad, Zeinab Khanafer, Michèle Prévost, Pierre Servais, Emile Sylvestre, and Sarah Dorner. 2021. "Automated Targeted Sampling of Waterborne Pathogens and Microbial Source Tracking Markers Using Near-Real Time Monitoring of Microbiological Water Quality" Water 13, no. 15: 2069. https://doi.org/10.3390/w13152069
APA StyleBurnet, J. -B., Habash, M., Hachad, M., Khanafer, Z., Prévost, M., Servais, P., Sylvestre, E., & Dorner, S. (2021). Automated Targeted Sampling of Waterborne Pathogens and Microbial Source Tracking Markers Using Near-Real Time Monitoring of Microbiological Water Quality. Water, 13(15), 2069. https://doi.org/10.3390/w13152069