Changes in Wastewater Treatment Performance and the Microbial Community during the Bioaugmentation of a Denitrifying Pseudomonas Strain in the Low Carbon–Nitrogen Ratio Sequencing Batch Reactor
Abstract
:1. Introduction
2. Materials and Methods
2.1. Media and Isolation Strategy
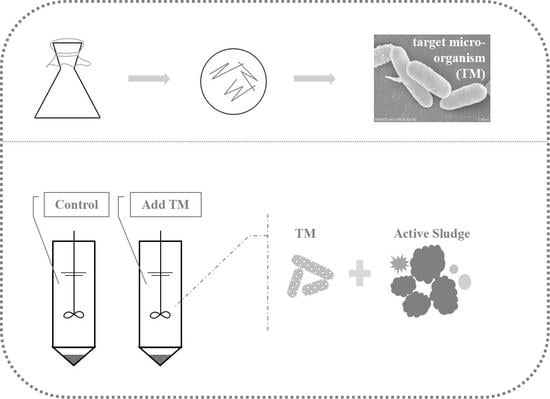
2.2. Batch Experiment
2.3. Bioreactor Operation Strategy
2.4. Analytical Methods
2.5. Microbial Community Analysis
3. Results
3.1. Isolation of Nitrogen Removal Bacteria
3.2. Denitrification in the Batch Study
3.3. Denitrification in SBR
3.4. Microbial Community Analysis in Lab-Scale SBR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Jin, Y.; Ding, D.; Feng, C.; Tong, S.; Suemura, T.; Zhang, F. Performance of sequencing batch biofilm reactors with different control systems in treating synthetic municipal wastewater. Bioresour. Technol. 2012, 104, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, B.; Ye, L. The combined effects of COD/N ratio and nitrate recycling ratio on nitrogen and phosphorus removal in anaerobic/anoxic/aerobic (A2/O)-biological aerated filter (BAF) systems. Biochem. Eng. J. 2015, 93, 235–242. [Google Scholar] [CrossRef]
- Loperena, L.; Ferrari, M.D.; Saravia, V.; Murro, D.; Lima, C.; Ferrando, L.; Fernandez, A.; Lareo, C. Performance of a commercial inoculum for the aerobic biodegradation of a high fat content dairy wastewater. Bioresour. Technol. 2007, 98, 1045–1051. [Google Scholar] [CrossRef] [PubMed]
- Bella, E.; Smith, P.G. The bioaugmentation of sequencing batch reactor sludges for biological phosphorus removal. Water Sci. Technol. 1997, 35, 19–26. [Google Scholar] [CrossRef]
- Oerther, D.B.; James, D.; Ebru, D.; Eric, L.; Freedman, D.L.; Lutgarde, R. Bioaugmentation of sequencing batch reactors for biological phosphorus removal: Comparative rrna sequence analysis and hybridization with oligonucleotide probes. Water Sci. Technol. 1998, 37, 469–473. [Google Scholar] [CrossRef]
- Singer, A.C.; van der Gast, C.J.; Thompson, I.P. Perspectives and vision for strain selection in bioaugmentation. Trends Biotechnol. 2005, 23, 74–77. [Google Scholar] [CrossRef]
- Boon, N.; Top, E.M.; Verstraete, W.; Siciliano, S.D. Bioaugmentation as a tool to protect the structure and function of an activated-sludge microbial community against a 3-chloroaniline shock load. Appl. Environ. Microbiol. 2003, 69, 1511–1520. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Liu, Y.; Wang, J.; Zhang, Y.; Yang, H. Influence of growth manner on nitrifying bacterial communities and nitrification kinetics in three lab-scale bioreactors. J. Ind. Microbiol. Biotechnol. 2012, 39, 595–604. [Google Scholar] [CrossRef]
- Zumft, W.G. Microbiology and Molecular Biology Reviews. Microbiol. Mol. Biol. Rev. 1997, 61, 533–616. [Google Scholar]
- Song, Z.; Zhang, X.; Sun, F.; Ngo, H.H.; Guo, W.; Wen, H.; Li, C.; Zhang, Z. Specific microbial diversity and functional gene (AOB amoA) analysis of a sponge-based aerobic nitrifying moving bed biofilm reactor exposed to typical pharmaceuticals. Sci. Total Environ. 2020, 742, 140660. [Google Scholar] [CrossRef]
- Pelaz, L.; Gomez, A.; Letona, A.; Garralon, G.; Fdz-Polanco, M. Nitrogen removal in domestic wastewater. Effect of nitrate recycling and COD/N ratio. Chemosphere 2018, 212, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, T.; Akiyama, T.; Fukumori, Y. Structure and function of NO reductase with oxygen-reducing activity. In Oxygen Homeostasis and Its Dynamics. Keio University Symposia for Life Science and Medicine; Ishimura, Y., Shimada, H., Suematsu, M., Eds.; Springer: Tokyo, Japan, 1998; Volume 1. [Google Scholar]
- Zhang, Q.L.; Liu, Y.; Ai, G.M.; Miao, L.L.; Zheng, H.Y.; Liu, Z.P. The characteristics of a novel heterotrophic nitrification-aerobic denitrification bacterium, Bacillus methylotrophicus strain L7. Bioresour. Technol. 2012, 108, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Joo, H.S.; Hirai, M.; Shoda, M. Characteristics of ammonium removal by heterotrophic nitrification-aerobic denitrification by Alcaligenes faecalis No. 4. J. Biosci. Bioeng. 2005, 100, 184–191. [Google Scholar] [CrossRef]
- Rijn, J.V.; Tal, Y.; Schreier, H.J. Denitrification in recirculating systems: Theory and applications. Aquac. Eng. 2006, 34, 364–376. [Google Scholar] [CrossRef]
- Yuan, H.; Huang, S.; Yuan, J.; You, Y.; Zhang, Y. Characteristics of microbial denitrification under different aeration intensities: Performance, mechanism, and co-occurrence network. Sci. Total Environ. 2020, 754, 141965. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Ni, J.; Ma, T.; Liu, T.; Zheng, M. Bioaugmentation treatment of municipal wastewater with heterotrophic-aerobic nitrogen removal bacteria in a pilot-scale SBR. Bioresour. Technol. 2015, 183, 25–32. [Google Scholar] [CrossRef]
- Layeghifard, M.; DM Hwang, D.M.; Guttman, D.S. Disentangling interactions in the microbiome: A network perspective. Trends Microbiol. 2017, 25, 217–228. [Google Scholar] [CrossRef]
- Takaya, N.; Catalan-Sakairi, M.A.B.; Sakaguchi, Y.; Kato, I.; Zhou, Z.; Shoun, H. Aerobic denitrifying bacteria that produce low levels of nitrous oxide. Appl. Environ. 2003, 69, 3152–3157. [Google Scholar] [CrossRef] [Green Version]
- Miranda, K.M.; Espey, M.G.; Wink, D.A. A rapid, simple spectrophotometric method for simultaneous detection of nitrate and nitrite. Nitric Oxide 2001, 5, 62–71. [Google Scholar] [CrossRef]
- Braker, G.; Tiedje, J.M. Nitric oxide reductase (norB) genes from pure cultures and environmental samples. Appl. Environ. 2003, 69, 3476–3483. [Google Scholar] [CrossRef] [Green Version]
- Throback, I.N.; Enwall, K.; Jarvis, A.; Hallin, S. Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol. Ecol. 2004, 49, 401–417. [Google Scholar] [CrossRef] [PubMed]
- Henry, S.; Bru, D.; Stres, B.; Hallet, S.; Philippot, L. Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl. Environ. 2006, 72, 5181–5189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, A.; Li, S.; Zhang, L.; Wang, H.; Yang, J.; Luo, Z.; Rashid, A.; Chen, S.; Huang, W.; Yu, C.P. Prokaryotic footprints in urban water ecosystems: A case study of urban landscape ponds in a coastal city, China. Environ. Pollut. 2018, 242 Pt B, 1729–1739. [Google Scholar] [CrossRef]
- Hildebrand, F.; T, R.; Voigt, A.Y.; Bork, P.; Raes, J. LotuS: An efficient and user-friendly OTU processing pipeline. Microbiome 2014, 2, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, H.H.H.; Wagner, H. Vegan: Community Ecology Package. R Package Version 2.5-6. 2019. Available online: http://cran.rproject.org/package=vegan (accessed on 28 December 2021).
- Kembel, S.W.; Cowan, P.D.; Helmus, M.R.; Cornwell, W.K.; Morlon, H.; Ackerly, D.D.; Blomberg, S.P.; Webb, C.O. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 2010, 26, 1463–1464. [Google Scholar] [CrossRef] [Green Version]
- Nepusz, G.C.T. The igraph software package for complex network research. Int. J. Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Harrell, F.E., Jr. Hmisc: Harrell Miscellaneous, R Package Version 4.4–0. 2020. Available online: https://cran.r-project.org/package=Hmisc (accessed on 28 December 2021).
- He, T.; Li, Z.; Sun, Q.; Xu, Y.; Ye, Q. Heterotrophic nitrification and aerobic denitrification by Pseudomonas tolaasii Y-11 without nitrite accumulation during nitrogen conversion. Bioresour. Technol. 2016, 200, 493–499. [Google Scholar] [CrossRef]
- Van Niel, E.W.J.; Braber, K.J.; Robertson, L.A.; Kuenen, J.G. Heterotrophic nitrification and aerobic denitrification in Alcaligenes faecalis strain TUD. Anton Leeuw. Int. J. Gen. 1992, 62, 231–237. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Q.; Li, M.; Sang, W.; Wang, Y.; Wu, L.; Yang, Y. Bioaugmentation of sequencing batch reactor for aniline treatment during start-up period: Investigation of microbial community structure of activated sludge. Chemosphere 2020, 243, 125426. [Google Scholar] [CrossRef]
- Chen, C.; Ali, A.; Su, J.; Wang, Y.; Huang, T.; Gao, J. Pseudomonas stutzeri GF2 augmented the denitrification of low carbon to nitrogen ratio: Possibility for sewage wastewater treatment. Bioresour. Technol. 2021, 333, 125–169. [Google Scholar] [CrossRef]
- Bouchez, T.; Patureau, D.; Delgenes, J.P.; Moletta, R. Successful bacterial incorporation into activated sludge flocs using alginate. Bioresour. Technol. 2009, 100, 1031–1032. [Google Scholar] [CrossRef]
- Boon, N.; De Gelder, L.; Lievens, H.; Siciliano, S.D.; Top, E.M.; Verstraete, W. Bioaugmenting bioreactors for the continuous removal of 3-chloroaniline by a slow release approach. Environ. Sci. Technol. 2002, 36, 4698–4704. [Google Scholar] [CrossRef]
- Bouchez, T.; Patureau, D.; Dabert, P.; Juretschko, S.; Doré, J.; Delgenès, P.; Moletta, R.; Wagner, M. Ecological study of a bioaugmentation failure. Environ. Microbiol. 2000, 2, 179–190. [Google Scholar] [CrossRef]
- Goldstein, R.M.; Mallory, L.M.; Alexander, M. Reasons for possible failure of inoculation to enhance biodegradation. Appl. Environ. Microbiol. 1985, 50, 977–983. [Google Scholar] [CrossRef] [Green Version]
- Bai, Y.; Sun, Q.; Sun, R.; Wen, D.; Tang, X. Bioaugmentation and adsorption treatment of coking wastewater containing pyridine and quinoline using zeolite-biological aerated filters. Environ. Sci. Technol. 2011, 45, 1940–1948. [Google Scholar] [CrossRef]
- Munakata-Marr, J.; Matheson, V.G.; Forney, L.J.; Tiedje, J.M.; McCarty, P.L. Long-term biodegradation of trichloroethylene influenced by bioaugmentation and dissolved oxygen in aquifer microcosms. Environ. Sci. Technol. 1997, 31, 786–791. [Google Scholar] [CrossRef]









Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, T.; Yang, X.; Sun, Q.; Hu, A.; Qin, D.; Li, J.; Wang, Y.; Yu, C.-P. Changes in Wastewater Treatment Performance and the Microbial Community during the Bioaugmentation of a Denitrifying Pseudomonas Strain in the Low Carbon–Nitrogen Ratio Sequencing Batch Reactor. Water 2022, 14, 540. https://doi.org/10.3390/w14040540
Chen T, Yang X, Sun Q, Hu A, Qin D, Li J, Wang Y, Yu C-P. Changes in Wastewater Treatment Performance and the Microbial Community during the Bioaugmentation of a Denitrifying Pseudomonas Strain in the Low Carbon–Nitrogen Ratio Sequencing Batch Reactor. Water. 2022; 14(4):540. https://doi.org/10.3390/w14040540
Chicago/Turabian StyleChen, Tianyuan, Xiaoyong Yang, Qian Sun, Anyi Hu, Dan Qin, Jiangwei Li, Yinhan Wang, and Chang-Ping Yu. 2022. "Changes in Wastewater Treatment Performance and the Microbial Community during the Bioaugmentation of a Denitrifying Pseudomonas Strain in the Low Carbon–Nitrogen Ratio Sequencing Batch Reactor" Water 14, no. 4: 540. https://doi.org/10.3390/w14040540
APA StyleChen, T., Yang, X., Sun, Q., Hu, A., Qin, D., Li, J., Wang, Y., & Yu, C. -P. (2022). Changes in Wastewater Treatment Performance and the Microbial Community during the Bioaugmentation of a Denitrifying Pseudomonas Strain in the Low Carbon–Nitrogen Ratio Sequencing Batch Reactor. Water, 14(4), 540. https://doi.org/10.3390/w14040540