3D-MRI Brain Tumor Detection Model Using Modified Version of Level Set Segmentation Based on Dragonfly Algorithm
Abstract
:1. Introduction
1.1. Problem Statement and Motivation
1.2. Contribution and Methodology
2. Related Work
3. Materials and Methods
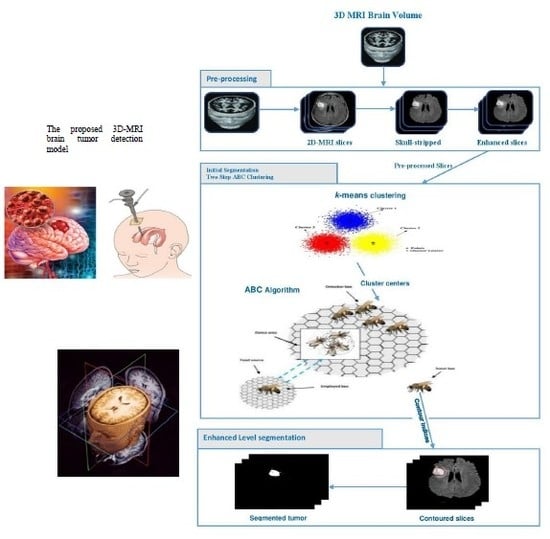
3.1. Preprocessing Phase
3.2. Two Steps Dragonfly-Based Clustering Phase
| Algorithm 1: Two-step Dragonfly Clustering. |
| Input: dataset contains MRI brain images Output: Best solution of final cluster center () Begin Initialization phase Initialize the position of dragonfly population Xi (i = 1 2, ..., n). Initialize step vectors Δ Xi For /* is the total number of food sources (number of clusters) */ Initialize the food source within the boundary of given dataset in random order; Evaluate the better potions of food sources by applying the k-means algorithm / *Algorithm 2*/ Send the dragonflies to the food sources; / * Computed centers */ End For Dragonfly algorithm Phase Iteration = 0; Do While (the end condition is not satisfied) For i = 1:n Calculate the fitness of each dragonfly Update the food source and enemy Update w, s, a, c, f, and e Calculate S, A, C, F, and E using Equations (4) to (8) Update neighboring radius If (a dragonfly has at least one neighboring dragonfly) Update step vector (ΔX) using Equation (9) Update position vector X using Equation (10) Else Update position vector using Equation (11) End if Check and correct the new positions based on the boundaries of variables End For For Compute the probability. /* Calculate the probability for each one */ End For For If (rand ( ) < ) /* denotes the probability associated with food source */ Calculate the new fitness of the new food source using Equation (14); Select the best food source by using a greedy selection between the old and new food source; Else ; End If End For End While Output: Final clusters‘ centers. End |
| Algorithm 2: K-means clustering [42]. |
| Input: . // the number of clusters; dataset contains MRI brain images (2D slices). Begin Arbitrary choose objects from as the initial cluster centers; Repeat - (re) group the most similar objects into a cluster, based on the Euclidian distance between the object and the cluster centroid (mean); - Update the cluster centroid, i.e., calculate the mean value of the objects for each cluster. Until no change. |
3.3. Level Set Segmentation
| Algorithm 3: Level set segmentation. |
| 1: Insert initial contour points using two-step DA clustering output (ROI indexes). 2: Construct a signed distance function. 3: Calculate feature image using Gaussian filter and gradient. 4: Obtain the curve’s narrow band. 5: Obtain curvature and use gradient descent to minimize energy. 6: Evolve the curve. 7: Repeat step number two and stop after obtaining the segmented region. |
4. Experimental Results
4.1. Experiment 1: Comparison with Existing Methods
4.2. Experiment 2: Model Accuracy with and without k-Means
4.3. Experiment 3: Role of DA to Reduce Level Set Iteration
5. Conclusions and Future Work
Author Contributions
Funding
Conflicts of Interest
References
- El-Melegy, M.T.; El-Magd, K.M.A.; Ali, S.A.; Hussain, K.F.; Mahdy, Y.B. Ensemble of Multiple Classifiers for Automatic Multimodal Brain Tumor Segmentation. In Proceedings of the International Conference on Innovative Trends in Computer Engineering (ITCE), Aswan, Egypt, 2–4 February 2019; pp. 58–63. [Google Scholar]
- Aparna, R.M.; Shanmugavadivu, P. A Survey of Medical Imaging, Storage and Transfer Techniques. In Proceedings of the International Conference on ISMAC in Computational Vision and Bio-Engineering, Coimbatore, India, 16–17 May 2018; pp. 17–29. [Google Scholar]
- Lu, C.; Xu, Z.; Ye, X. Evaluation of Intraoperative MRI-Assisted Stereotactic Brain Tissue Biopsy: A Single-Center Experience in China. Chin. Neurosurg. J. 2019, 5, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Bauer, S.; Wiest, R.; Nolte-P, L.; Reyes, M. A Survey of MRI-Based Medical Image Analysis For Brain Tumor Studies. Phys. Med. Biol. 2013, 58, 1–44. [Google Scholar] [CrossRef] [PubMed]
- Mild, K.H.; Lundström, R.H.; Wilén, J.H. Non-ionizing Radiation in Swedish Health Care Exposure and Safety Aspects. Int. J. Environ. Res. Public Health 2019, 16, 1186. [Google Scholar] [CrossRef] [Green Version]
- Zhuge, Y.; Krauze, A.V.; Ning, H.; Cheng, J.Y.; Arora, B.C.; Camphausen, K.; Miller, R.W. Brain tumor segmentation using holistically nested neural networks in MRI images. Med. Phys. 2017, 44, 5234–5243. [Google Scholar] [CrossRef]
- Banerjee, S.; Mitra, S. Novel Volumetric Sub-region Segmentation in Brain Tumors. Front. Comput. Neurosci. 2020, 14, 1–13. [Google Scholar] [CrossRef]
- Popoola, J.J.; Godson, T.E.; Olasoji, Y.O.; Adu, M.R. Study on Capabilities of Different Segmentation Algorithms in Detecting and Reducing Brain Tumor Size in Magnetic Resonance Imaging for Effective Telemedicine Services. Eur. J. Eng. Res. Sci. 2019, 4, 23–29. [Google Scholar] [CrossRef]
- Angulakshmi, M.; Priya, G.L. Automated Brain Tumor Segmentation Techniques—A Review. Int. J. Imaging Syst. Technol. 2017, 27, 66–77. [Google Scholar] [CrossRef] [Green Version]
- Shirly, S.; Ramesh, K. Review on 2D and 3D MRI Image Segmentation Techniques. Curr. Med. Imaging Former. Curr. Med. Imaging Rev. 2019, 15, 150–160. [Google Scholar] [CrossRef]
- Sajid, S.; Hussain, S.; Sarwar, A. Brain Tumor Detection and Segmentation in MR Images Using Deep Learning. Arab. J. Sci. Eng. 2019, 44, 9249–9261. [Google Scholar] [CrossRef]
- Wang, D. Efficient Level-Set Segmentation Model Driven by The Local GMM and Split Bregman Method. IET Image Process. 2019, 13, 761–770. [Google Scholar] [CrossRef]
- Rahman, C.; Rashid, T. Dragonfly Algorithm and Its Applications in Applied Science Survey. Comput. Intell. Neurosci. 2019, 2019, 11–21. [Google Scholar] [CrossRef]
- Seyedali, M. Dragonfly Algorithm: A New Meta-Heuristic Optimization Technique for Solving Single-Objective, Discrete, and Multi-Objective Problems. Neural Comput. Appl. 2016, 27, 1053–1073. [Google Scholar] [CrossRef]
- Ranjini, K.; Murugan, S. Memory Based Hybrid Dragonfly Algorithm for Numerical Optimization Problems. Expert Syst. Appl. 2017, 83, 63–78. [Google Scholar] [CrossRef]
- Saman, S.; Narayanan, S.J. Survey on brain tumor segmentation and feature extraction of MR images. Int. J. Multimed. Inf. Retr. 2018, 8, 79–99. [Google Scholar] [CrossRef]
- Tiwari, A.; Srivastava, S.; Pant, M. Brain tumor segmentation and classification from magnetic resonance images: Review of selected methods from 2014 to 2019. Pattern Recognit. Lett. 2020, 131, 244–260. [Google Scholar] [CrossRef]
- El-Baz, A.; Suri, J.S. Level Set Method in Medical Imaging Segmentation; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- Al-Rifaie, M.; Aber, A.; Hemanth, J. Deploying Swarm Intelligence in Medical Imaging; Identifying Metastasis, Micro-Calcifications and Brain Image Segmentation. IET Syst. Biol. 2015, 9, 234–244. [Google Scholar] [CrossRef]
- Rupika, N.; Menon, H.; Vikram, K. A Survey on Advanced Segmentation Techniques for Brain MRI Image Segmentation. Int. J. Adv. Sci. Eng. Inf. Technol. 2017, 7, 1448–1456. [Google Scholar] [CrossRef] [Green Version]
- Kermi, A.; Andjouh, K.; Zidane, F. Fully automated brain tumor segmentation system in 3D-MRI using symmetry analysis of brain and level-sets. IET Image Process. 2018, 12, 1964–1971. [Google Scholar] [CrossRef]
- Bal, A.; Banerjee, M.; Chakrabarti, A.; Sharma, P. MRI Brain Tumor Segmentation and Analysis using Rough-Fuzzy C-Means and Shape Based Properties. J. King Saud Univ. Comput. Inf. Sci. 2018. [Google Scholar] [CrossRef]
- Anitha, V.; Murugavalli, S. Brain Tumor Classification using Two-Tier Classifier with Adaptive Segmentation Technique. IET Comput. Vis. 2016, 10, 9–17. [Google Scholar] [CrossRef]
- Mahalakshmi, A.; Krishnappa, H.; Jayadevappa, D. A Hybrid Approach for the Segmentation of Brain Tumor using K-Means Clustering and Variational Level Set. J. Adv. Res. Dyn. Control. Syst. 2018, 10, 258–264. [Google Scholar]
- Thapaliya, K.; Pyun, J.-Y.; Park, C.-S.; Kwon, G.-R. Level Set Method with Automatic Selective Local Statistics for Brain Tumor Segmentation in MR Images. Comput. Med. Imaging Graph. 2013, 37, 522–537. [Google Scholar] [CrossRef] [PubMed]
- Le, T.; Gummadi, R.; Savvides, M. Deep Recurrent Level Set for Segmenting Brain Tumors. Lect. Notes Comput. Sci. 2018, 11072, 646–653. [Google Scholar] [CrossRef] [Green Version]
- Qin, P.; Zhang, J.; Zeng, J.; Liu, H.; Cui, Y. A Framework Combining DNN and Level-Set Method to Segment Brain Tumor in Multi-Modalities MR Image. Soft Comput. 2019, 19, 9237–9251. [Google Scholar] [CrossRef]
- Rajan, P.G.; Sundar, C. Brain Tumor Detection and Segmentation by Intensity Adjustment. J. Med. Syst. 2019, 43, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Maksoud, E.; Elmogy, M.; Al-Awadi, R. Brain Tumor Segmentation Based on A Hybrid Clustering Technique. Egypt. Inform. J. 2015, 16, 71–81. [Google Scholar] [CrossRef]
- Ural, B. A Computer-Based Brain Tumor Detection Approach with Advanced Image Processing and Probabilistic Neural Network Methods. J. Med. Biol. Eng. 2017, 38, 867–879. [Google Scholar] [CrossRef]
- Ma, C.; Luo, G.; Wang, K. Concatenated and Connected Random Forests with Multiscale Patch Driven Active Contour Model for Automated Brain Tumor Segmentation of MR Images. IEEE Trans. Med. Imaging 2018, 37, 1943–1954. [Google Scholar] [CrossRef]
- Hancer, E.; Ozturk, C.; Karaboga, D. Extraction of Brain Tumors from MRI Images with Artificial Bee Colony-Based Segmentation Methodology. In Proceedings of the 8th International Conference on Electrical and Electronics Engineering (ELECO), Bursa, Turkey, 28–30 November 2019; pp. 516–520. [Google Scholar]
- Christ, J.; Subramanian, R. Clown Fish Queuing and Switching Optimization Algorithm for Brain Tumor Segmentation. Biomed. Res. 2016, 27, 65–69. [Google Scholar]
- Narayanan, B.; Hardie, R. A Computationally Efficient U-Net Architecture for Lung Segmentation in Chest Radiographs. In Proceedings of the IEEE National Aerospace and Electronics Conference (NAECON), Dayton, OH, USA, 20–24 July 2020; pp. 279–284. [Google Scholar]
- Jin, L.; Pan, Y.; Li, M.; Chen, Z.; Tang, L.; Lu, C.; Wang, J. Applications of Deep Learning to MRI Images: A Survey. Big Data Min. Anal. 2018, 1, 1–18. [Google Scholar] [CrossRef]
- Mohammad, H.; Davy, A.; Warde-Farley, D.; Biard, A.; Courville, A.; Bengio, Y.; Pal, C.; Jodoin, P.-M.; Larochelle, H. Brain Tumor Segmentation with Deep Neural Networks. Med. Image Anal. 2017, 35, 18–31. [Google Scholar] [CrossRef] [Green Version]
- Iglesias, J.E.; Liu, C.-Y.; Thompson, P.; Tu, Z. Robust brain extraction across datasets and comparison with publicly available methods. IEEE Trans. Med. Imaging 2011, 30, 1617–1634. [Google Scholar] [CrossRef] [PubMed]
- Nair, R.; David, E.; Rajagopal, S. A robust anisotropic diffusion filter with low arithmetic complexity for images. EURASIP J. Image Video Process. 2019, 48, 1–14. [Google Scholar] [CrossRef]
- Chen, J.; Gong, Y. Particle swarm optimization for two-echelon location-routing problem. J. Comput. Appl. 2013, 33, 2261–2264. [Google Scholar] [CrossRef]
- Kumar, Y.; Sahoo, G. A two-step artificial bee colony algorithm for clustering. Neural Comput. Appl. 2015, 28, 537–551. [Google Scholar] [CrossRef]
- Zhang, C.; Shen, X.; Cheng, H.; Qian, Q. Brain tumor segmentation based on hybrid clustering and morphological operations. Int. J. Biomed. Imaging 2019, 2019, 1–111. [Google Scholar] [CrossRef]
- Armano, G.; Farmani, M.R. Clustering analysis with combination of artificial bee colony algorithm and k-means technique. Int. J. Comput. Theory Eng. 2014, 6, 141–145. [Google Scholar] [CrossRef] [Green Version]
- Arunprasath, S. Internet of medical things-load optimization of power flow based on hybrid enhanced grey wolf optimization and dragonfly algorithm. Future Gener. Comput. Syst. 2019, 98, 319–330. [Google Scholar] [CrossRef]
- Bao, X.; Jia, H.; Lang, C. Dragonfly algorithm with opposition-based learning for multilevel thresholding Color Image Segmentation. Symmetry 2019, 11, 716. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Xu, C.; Gui, C.; Fox, M.D. Distance regularized level set evolution and its application to image segmentation. IEEE Trans. Image Process. 2010, 19, 3243–3254. [Google Scholar] [CrossRef]
- Belaid, A.; Boukerroui, D.; Maingourd, Y.; Lerallut, J.F. Phase-based level set segmentation of ultrasound images. IEEE Trans. Inf. Technol. Biomed. 2010, 15, 138–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menze, B.; Jakab, A.; Bauer, S.; Cramer, J.K.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 2015, 34, 1993–2024. [Google Scholar] [CrossRef] [PubMed]
- James, J.A.; Dasarathy, B. Medical image fusion: A survey of the state of the art. Int. J. Inf. Fusion 2014, 19, 4–19. [Google Scholar] [CrossRef] [Green Version]
- Lefkovits, L.; Lefkovits, S.; Vaida, M. Brain tumor segmentation based on random forest. Mem. Sci. Sect. Rom. Acad. 2016, 39, 83–93. [Google Scholar]
- Ayachi, R.; Amor, N.B. Brain tumor segmentation using support vector machines. Lect. Notes Comput. Sci. Book Ser. (LNCS) 2009, 5590, 736–747. [Google Scholar] [CrossRef]
- Zhang, K.; Zhang, L.; Song, H.; Zhou, W. Active contours with selective local or global segmentation: A new formulation and level set method. Image Vis. Comput. 2010, 28, 668–676. [Google Scholar] [CrossRef]











| Methods | Accuracy | Recall | Precision |
|---|---|---|---|
| Proposed Model (Two-step DA, Level Set) | 98.20 | 95.13 | 93.21 |
| Symmetry Analysis, Level Set [21] | 93.63 | 89.10 | 90.45 |
| Fuzzy C-Means [22] | 85.7 | 87.6 | 72.3 |
| Rough Fuzzy C-Means [22] | 91.50 | 90 | 92 |
| K-means, Level Set [24] | 89.30 | 92.7 | 75.8 |
| Random Forest [49] | 85.60 | 91.85 | 78.3 |
| Support Vector Machine (SVM) [50] | 94.25 | 92.15 | 91.21 |
| DNN Methods | Accuracy | Recall | Precision |
|---|---|---|---|
| Proposed Model (Two-step DA, Level Set) | 98.15 | 95.40 | 93.57 |
| Two-pathway CNN [36] | 96.24 | 89.67 | 82.56 |
| DNN, level set [26] | 91.58 | 96.40 | 93.23 |
| Nature-Inspired Metaheuristic | Accuracy | Recall | Precision |
|---|---|---|---|
| DA, Level Set | 98.15 | 95.40 | 93.57 |
| ABC, Level Set | 95.90 | 92.13 | 91.40 |
| PSO, Level Set | 93.58 | 92.40 | 89.23 |
| CF, Level Set | 96.85 | 94.32 | 92.55 |
| Methods | Accuracy | Mean | Standard Deviation |
|---|---|---|---|
| k-means, DA and level set | 98.10 | 95.67 | 0.02 |
| DA, level set | 85.67 | 82.56 | 0.04 |
| Methods | Patient No.1 | Patient No.2 | Patient No.3 | Patient No.4 | Patient No.5 |
|---|---|---|---|---|---|
| Level set with DA clustering | 15 | 18 | 16 | 15 | 20 |
| Level set without DA clustering | 252 | 330 | 371 | 266 | 407 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khalil, H.A.; Darwish, S.; Ibrahim, Y.M.; Hassan, O.F. 3D-MRI Brain Tumor Detection Model Using Modified Version of Level Set Segmentation Based on Dragonfly Algorithm. Symmetry 2020, 12, 1256. https://doi.org/10.3390/sym12081256
Khalil HA, Darwish S, Ibrahim YM, Hassan OF. 3D-MRI Brain Tumor Detection Model Using Modified Version of Level Set Segmentation Based on Dragonfly Algorithm. Symmetry. 2020; 12(8):1256. https://doi.org/10.3390/sym12081256
Chicago/Turabian StyleKhalil, Hassan A., Saad Darwish, Yasmine M. Ibrahim, and Osama F. Hassan. 2020. "3D-MRI Brain Tumor Detection Model Using Modified Version of Level Set Segmentation Based on Dragonfly Algorithm" Symmetry 12, no. 8: 1256. https://doi.org/10.3390/sym12081256
APA StyleKhalil, H. A., Darwish, S., Ibrahim, Y. M., & Hassan, O. F. (2020). 3D-MRI Brain Tumor Detection Model Using Modified Version of Level Set Segmentation Based on Dragonfly Algorithm. Symmetry, 12(8), 1256. https://doi.org/10.3390/sym12081256