Influence of Cell Type in In Vitro Induced Reprogramming in Cattle
Abstract
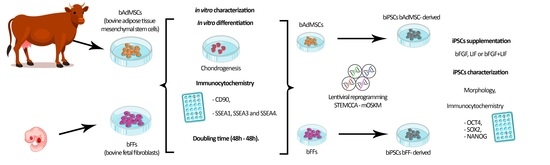
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Mesodermal In Vitro Differentiation
2.3. Doubling Time Analysis
2.4. Immunocytochemistry for CD90, SSEA1, 3 and 4
2.5. Lentiviral Production and Cellular Reprogramming
2.6. Characterization of biPSCs-Immunophenotyping
3. Results
3.1. bFFs and bAdMSCs Analysis
3.2. Doubling Time Analysis
3.3. Reprogramming Efficiency and Morphological Analysis of iPSCs
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Telfer, E.E.; Anderson, R.A. The Existence and Potential of Germline Stem Cells in the Adult Mammalian Ovary. Climacteric 2019, 22, 22–26. [Google Scholar] [CrossRef]
- Pieri, N.C.G.; de Souza, A.F.; Botigelli, R.C.; Machado, L.S.; Ambrosio, C.E.; dos Santos Martins, D.; de Andrade, A.F.C.; Meirelles, F.V.; Hyttel, P.; Bressan, F.F. Stem Cells on Regenerative and Reproductive Science in Domestic Animals. Vet. Res. Commun. 2019, 43, 7–16. [Google Scholar] [CrossRef] [PubMed]
- De Paepe, C.; Krivega, M.; Cauffman, G.; Geens, M.; Van de Velde, H. Totipotency and Lineage Segregation in the Human Embryo. Mol. Hum. Reprod. 2014, 20, 599–618. [Google Scholar] [CrossRef] [Green Version]
- Cao, H.; Yang, P.; Pu, Y.; Sun, X.; Yin, H.; Zhang, Y.; Zhang, Y.; Li, Y.; Liu, Y.; Fang, F.; et al. Characterization of Bovine Induced Pluripotent Stem Cells by Lentiviral Transduction of Reprogramming Factor Fusion Proteins. Int. J. Biol. Sci. 2012, 8, 498–511. [Google Scholar] [CrossRef] [PubMed]
- Goszczynski, D.E.; Cheng, H.; Demyda-Peyrás, S.; Medrano, J.F.; Wu, J.; Ross, P.J. In Vitro Breeding: Application of Embryonic Stem Cells to Animal Production. Biol. Reprod. 2019, 100, 885–895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sampaio, R.V.; Chiaratti, M.R.; Santos, D.C.N.; Bressan, F.F.; Sangalli, J.R.; Sá, A.L.A.; Silva, T.V.G.; Costa, N.N.; Cordeiro, M.S.; Santos, S.S.D.; et al. Generation of Bovine (Bos indicus) and Buffalo (Bubalus bubalis) Adipose Tissue Derived Stem Cells: Isolation, Characterization, and Multipotentiality. Genet. Mol. Res. 2015, 14, 53–62. [Google Scholar] [CrossRef]
- Lee, J.; Byeon, J.S.; Gu, N.Y.; Lee, S.; Lee, S.A.; Jeong, D.U.; Ouh, I.O.; Cho, I.S.; Song, J.Y.; Lee, Y.H.; et al. Bovine Tongue Epithelium-Derived Cells: A New Source of Bovine Mesenchymal Stem Cells. Biosci. Rep. 2020, 40, BSR20181829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sundaravadivelu, P.K.; Raina, K.; Thool, M.; Ray, A.; Joshi, J.M.; Kaveeshwar, V.; Sudhagar, S.; Lenka, N.; Thummer, R.P. Tissue-restricted stem cells as starting cell source for efficient generation of pluripotent stem cells: An overview. In Cell Biology and Translational Medicine, Volume 15: Stem Cells in Tissue Differentiation, Regulation and Disease; Turksen, K., Ed.; Springer International Publishing: Cham, Switzerland, 2022; pp. 151–180. ISBN 978-3-031-02378-1. [Google Scholar]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Machado, L.S.; Pieri, N.C.G.; Botigelli, R.C.; de Castro, R.V.G.; de Souza, A.F.; Bridi, A.; Lima, M.A.; Fantinato Neto, P.; de Figueiredo Pessôa, L.V.; Martins, S.M.M.K.; et al. Generation of Neural Progenitor Cells (NPC) from Porcine Induced Pluripotent Stem Cells (PiPSC). J. Tissue Eng. Regen. Med. 2020, 14, 1880–1891. [Google Scholar] [CrossRef] [PubMed]
- Pieri, N.C.G.; de Souza, A.F.; Botigelli, R.C.; Pessôa, L.V.d.F.; Recchia, K.; Machado, L.S.; Glória, M.H.; de Castro, R.V.G.; Leal, D.F.; Fantinato Neto, P.; et al. Porcine Primordial Germ Cell-Like Cells Generated from Induced Pluripotent Stem Cells Under Different Culture Conditions. Stem Cell Rev. Rep. 2022, 18, 1639–1656. [Google Scholar] [CrossRef]
- West, F.D.; Terlouw, S.L.; Kwon, D.J.; Mumaw, J.L.; Dhara, S.K.; Hasneen, K.; Dobrinsky, J.R.; Stice, S.L. Porcine Induced Pluripotent Stem Cells Produce Chimeric Offspring. Stem Cells Dev. 2010, 19, 1211–1220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Secher, J.; Hyttel, P.; Ivask, M.; Kolko, M.; Hall, V.J.; Freude, K.K. Generation of Transgene-Free Porcine Intermediate Type Induced Pluripotent Stem Cells. Cell Cycle 2018, 17, 2547–2563. [Google Scholar] [CrossRef] [Green Version]
- Pessôa, L.V.d.F.; Pires, P.R.L.; del Collado, M.; Pieri, N.C.G.; Recchia, K.; Souza, A.F.; Perecin, F.; da Silveira, J.C.; de Andrade, A.F.C.; Ambrosio, C.E.; et al. Generation and MiRNA Characterization of Equine Induced Pluripotent Stem Cells Derived from Fetal and Adult Multipotent Tissues. Stem Cells Int. 2019, 2019, 1393791. [Google Scholar] [CrossRef] [Green Version]
- De Castro, R.V.G.; Pieri, N.C.G.; Fantinato Neto, P.; Grizendi, B.M.; Dória, R.G.S.; Meirelles, F.V.; Smith, L.C.; Garcia, J.M.; Bressan, F.F. In Vitro Induction of Pluripotency from Equine Fibroblasts in 20% or 5% Oxygen. Stem Cells Int. 2020, 2020, 8814989. [Google Scholar] [CrossRef]
- Deng, Y.; Liu, Q.; Luo, C.; Chen, S.; Li, X.; Wang, C.; Liu, Z.; Lei, X.; Zhang, H.; Sun, H.; et al. Generation of Induced Pluripotent Stem Cells from Buffalo (Bubalus bubalis) Fetal Fibroblasts with Buffalo Defined Factors. Stem Cells Dev. 2012, 21, 2485–2494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bressan, F.F.; Bassanezze, V.; de Figueiredo Pessôa, L.V.; Sacramento, C.B.; Malta, T.M.; Kashima, S.; Fantinato Neto, P.; Strefezzi, R.D.F.; Pieri, N.C.G.; Krieger, J.E.; et al. Generation of Induced Pluripotent Stem Cells from Large Domestic Animals. Stem Cell Res. Ther. 2020, 11, 247. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; An, X.L.; Yu, H.; Cai, N.N.; Zhai, Y.H.; Li, Q.; Cheng, H.; Zhang, S.; Tang, B.; Li, Z.Y.; et al. Transcriptome Profile of Bovine IPSCs Derived from Sertoli Cells. Theriogenology 2020, 146, 120–132. [Google Scholar] [CrossRef]
- Kawaguchi, T.; Tsukiyama, T.; Kimura, K.; Matsuyama, S.; Minami, N.; Yamada, M.; Imai, H. Generation of Naïve Bovine Induced Pluripotent Stem Cells Using Piggybac Transposition of Doxycycline-Inducible Transcription Factors. PLoS ONE 2015, 10, e0135403. [Google Scholar] [CrossRef] [Green Version]
- Bogliotti, Y.S.; Wu, J.; Vilarino, M.; Okamura, D.; Soto, D.A.; Zhong, C.; Sakurai, M.; Sampaio, R.V.; Suzuki, K.; Izpisua Belmonte, J.C.; et al. Efficient Derivation of Stable Primed Pluripotent Embryonic Stem Cells from Bovine Blastocysts. Proc. Natl. Acad. Sci. USA 2018, 115, 2090–2095. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, K.; Ohta, H.; Kurimoto, K.; Aramaki, S.; Saitou, M. Reconstitution of the Mouse Germ Cell Specification Pathway in Culture by Pluripotent Stem Cells. Cell 2011, 146, 519–532. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, K. Offspring from Oocytes Derived from in Vitro Primordial Germ Cell-like Cells in Mice. Science 2012, 971, 10–15. [Google Scholar] [CrossRef] [Green Version]
- Hikabe, O.; Hamazaki, N.; Nagamatsu, G.; Obata, Y.; Hirao, Y.; Hamada, N.; Shimamoto, S.; Imamura, T.; Nakashima, K.; Saitou, M.; et al. Reconstitution in Vitro of the Entire Cycle of the Mouse Female Germ Line. Nature 2016, 539, 299–303. [Google Scholar] [CrossRef] [PubMed]
- Canizo, J.R.; Vazquez Echegaray, C.; Klisch, D.; Aller, J.F.; Paz, D.A.; Alberio, R.H.; Alberio, R.; Guberman, A.S. Exogenous Human OKSM Factors Maintain Pluripotency Gene Expression of Bovine and Porcine IPS-like Cells Obtained with STEMCCA Delivery System. BMC Res. Notes 2018, 11, 509. [Google Scholar] [CrossRef] [Green Version]
- Han, X.; Han, J.; Ding, F.; Cao, S.; Lim, S.S.; Dai, Y.; Zhang, R.; Zhang, Y.; Lim, B.; Li, N. Generation of Induced Pluripotent Stem Cells from Bovine Embryonic Fibroblast Cells. Cell Res. 2011, 21, 1509–1512. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Wang, Z.; Zhang, J.; Yang, J.; Gao, X.; Wu, B.; Zhao, G.; Bao, S.; Hu, S.; Liu, P.; et al. Characterization of the Single-Cell Derived Bovine Induced Pluripotent Stem Cells. Tissue Cell 2017, 49, 521–527. [Google Scholar] [CrossRef]
- Cravero, D.; Martignani, E.; Miretti, S.; Accornero, P.; Pauciullo, A.; Sharma, R.; Donadeu, F.X.; Baratta, M. Generation of Induced Pluripotent Stem Cells from Bovine Epithelial Cells and Partial Redirection toward a Mammary Phenotype In Vitro. Cell. Reprogr. 2015, 17, 211–220. [Google Scholar] [CrossRef]
- Wang, S.W.; Wang, S.S.W.; Wu, D.C.; Lin, Y.C.; Ku, C.C.; Wu, C.C.; Chai, C.Y.; Lee, J.N.; Tsai, E.M.; Lin, C.L.S.; et al. Androgen Receptor-Mediated Apoptosis in Bovine Testicular Induced Pluripotent Stem Cells in Response to Phthalate Esters. Cell Death Dis. 2013, 4, e907. [Google Scholar] [CrossRef] [Green Version]
- Talluri, T.R.; Kumar, D.; Glage, S.; Garrels, W.; Ivics, Z.; Debowski, K.; Behr, R.; Niemann, H.; Kues, W.A. Derivation and Characterization of Bovine Induced Pluripotent Stem Cells by Transposon-Mediated Reprogramming. Cell. Reprogr. 2015, 17, 131–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Platero-Luengo, A.; Sakurai, M.; Sugawara, A.; Gil, M.A.; Yamauchi, T.; Suzuki, K.; Bogliotti, Y.S.; Cuello, C.; Morales Valencia, M.; et al. Interspecies Chimerism with Mammalian Pluripotent Stem Cells. Cell 2017, 168, 473–486.e15. [Google Scholar] [CrossRef] [Green Version]
- Raab, S.; Klingenstein, M.; Liebau, S.; Linta, L. A Comparative View on Human Somatic Cell Sources for IPSC Generation. Stem Cells Int. 2014, 2014, 768391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ray, A.; Joshi, J.M.; Sundaravadivelu, P.K.; Raina, K.; Lenka, N.; Kaveeshwar, V.; Thummer, R.P. An Overview on Promising Somatic Cell Sources Utilized for the Efficient Generation of Induced Pluripotent Stem Cells. Stem Cell Rev. Rep. 2021, 17, 1954–1974. [Google Scholar] [CrossRef] [PubMed]
- Pessôa, L.V.d.F.; Bressan, F.F.; Freude, K.K. Induced Pluripotent Stem Cells throughout the Animal Kingdom: Availability and Applications. World J. Stem Cells 2019, 11, 491–505. [Google Scholar] [CrossRef]
- Gruber, H.E.; Somayaji, S.; Riley, F.; Hoelscher, G.L.; Norton, H.J.; Ingram, J.; Hanley, E.N. Human Adipose-Derived Mesenchymal Stem Cells: Serial Passaging, Doubling Time and Cell Senescence. Biotech. Histochem. 2012, 87, 303–311. [Google Scholar] [CrossRef]
- Sumer, H.; Liu, J.; Malaver-Ortega, L.F.; Lim, M.L.; Khodadadi, K.; Verma, P.J. NANOG Is a Key Factor for Induction of Pluripotency in Bovine Adult Fibroblasts. J. Anim. Sci. 2011, 89, 2708–2716. [Google Scholar] [CrossRef] [Green Version]
- Czapla, J.; Matuszczak, S.; Kulik, K.; Wiśniewska, E.; Pilny, E.; Jarosz-Biej, M.; Smolarczyk, R.; Sirek, T.; Zembala, M.O.; Zembala, M.; et al. The Effect of Culture Media on Large-Scale Expansion and Characteristic of Adipose Tissue-Derived Mesenchymal Stromal Cells. Stem Cell Res. Ther. 2019, 10, 235. [Google Scholar] [CrossRef] [Green Version]
- Kolios, G.; Moodley, Y. Introduction to Stem Cells and Regenerative Medicine. Respiration 2012, 85, 3–10. [Google Scholar] [CrossRef]
- Zaim, M.; Karaman, S.; Cetin, G.; Isik, S. Donor Age and Long-Term Culture Affect Differentiation and Proliferation of Human Bone Marrow Mesenchymal Stem Cells. Ann. Hematol. 2012, 91, 1175–1186. [Google Scholar] [CrossRef]
- Kretlow, J.D.; Jin, Y.Q.; Liu, W.; Zhang, W.J.; Hong, T.H.; Zhou, G.; Baggett, L.S.; Mikos, A.G.; Cao, Y. Donor Age and Cell Passage Affects Differentiation Potential of Murine Bone Marrow-Derived Stem Cells. BMC Cell Biol. 2008, 9, 60. [Google Scholar] [CrossRef] [Green Version]
- Ruiz, S.; Panopoulos, A.D.; Herrerías, A.; Bissig, K.-D.; Lutz, M.; Berggren, W.T.; Verma, I.M.; Belmonte, J.C.I. A High Proliferation Rate Is Required for Cell Reprogramming and Maintenance of Human Embryonic Stem Cell Identity. Curr. Biol. 2011, 21, 45–52. [Google Scholar] [CrossRef] [Green Version]
- Bessi, B.W.; Botigelli, R.C.; Pieri, N.C.G.; Machado, L.S.; Cruz, J.B.; de Moraes, P.; de Souza, A.F.; Recchia, K.; Barbosa, G.; de Castro, R.V.G.; et al. Cattle In Vitro Induced Pluripotent Stem Cells Generated and Maintained in 5 or 20% Oxygen and Different Supplementation. Cells 2021, 10, 1531. [Google Scholar] [CrossRef] [PubMed]
- Nicola, N.A.; Babon, J.J. Leukemia Inhibitory Factor (LIF). Cytokine Growth Factor Rev. 2015, 26, 533–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, M. Therapeutics Targeting FGF Signaling Network in Human Diseases. Trends Pharmacol. Sci. 2016, 37, 1081–1096. [Google Scholar] [CrossRef]





| Total Colonies | AP Positive Colonies | AP Positive/Total % | |
|---|---|---|---|
| bFGF | 24 | 22 | 91.66% |
| LIF | 23 | 21 | 91.30% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Recchia, K.; Pessôa, L.V.d.F.; Pieri, N.C.G.; Pires, P.R.L.; Bressan, F.F. Influence of Cell Type in In Vitro Induced Reprogramming in Cattle. Life 2022, 12, 1139. https://doi.org/10.3390/life12081139
Recchia K, Pessôa LVdF, Pieri NCG, Pires PRL, Bressan FF. Influence of Cell Type in In Vitro Induced Reprogramming in Cattle. Life. 2022; 12(8):1139. https://doi.org/10.3390/life12081139
Chicago/Turabian StyleRecchia, Kaiana, Laís Vicari de Figueiredo Pessôa, Naira Caroline Godoy Pieri, Pedro Ratto Lisboa Pires, and Fabiana Fernandes Bressan. 2022. "Influence of Cell Type in In Vitro Induced Reprogramming in Cattle" Life 12, no. 8: 1139. https://doi.org/10.3390/life12081139
APA StyleRecchia, K., Pessôa, L. V. d. F., Pieri, N. C. G., Pires, P. R. L., & Bressan, F. F. (2022). Influence of Cell Type in In Vitro Induced Reprogramming in Cattle. Life, 12(8), 1139. https://doi.org/10.3390/life12081139