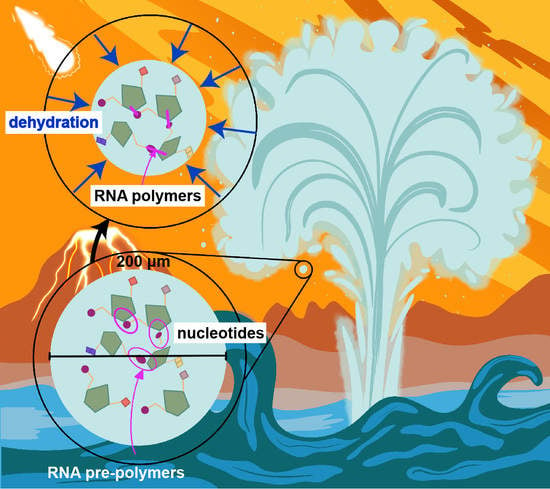
The Formation of RNA Pre-Polymers in the Presence of Different Prebiotic Mineral Surfaces Studied by Molecular Dynamics Simulations
Abstract
:1. Introduction
2. Results
2.1. Substrate Properties: Charge and Hydrogen Bond Potential
2.1.1. Charges
2.1.2. Hydrogen Bond Potential
2.2. Molecular Dynamics Simulations
2.3. Substrates with Positive Charges Attract the Nucleotides’ Phosphate Group
2.4. Substrates form Hydrogen Bonds with Different Parts of the Nucleotides
2.5. No Substrate Is a Good Catalyst for the Formation of Pre-Polymers
2.6. Substrate Surfaces Can Align Nucleotide through Interactions
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Substrates
5.2. Molecular Dynamics Simulations
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martin, W.; Baross, J.; Kelley, D.; Russell, M.J. Hydrothermal vents and the origin of life. Nat. Rev. Microbiol. 2008, 6, 805–814. [Google Scholar] [CrossRef] [PubMed]
- Wächtershäuser, G. Evolution of the first metabolic cycles. Proc. Natl. Acad. Sci. USA 1990, 87, 200–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sojo, V.; Herschy, B.; Whicher, A.; Camprubi, E.; Lane, N. The origin of life in alkaline hydrothermal vents. Astrobiology 2016, 16, 181–197. [Google Scholar] [CrossRef] [PubMed]
- Powner, M.W.; Gerland, B.; Sutherland, J.D. Synthesis of activated pyrimidine ribonucleotides in prebiotically plausible conditions. Nature 2009, 459, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Baross, J.A.; Hoffman, S.E. Submarine hydrothermal vents and associated gradient environments as sites for the origin and evolution of life. Orig. Life Evol. Biosph. 1985, 15, 327–345. [Google Scholar] [CrossRef]
- Ooka, H.; McGlynn, S.E.; Nakamura, R. Electrochemistry at deep-sea hydrothermal vents: Utilization of the thermodynamic driving force towards the autotrophic origin of life. ChemElectroChem 2019, 6, 1316–1323. [Google Scholar] [CrossRef] [Green Version]
- Macleod, G.; McKeown, C.; Hall, A.J.; Russell, M.J. Hydrothermal and oceanic pH conditions of possible relevance to the origin of life. Orig. Life Evol. Biosph. 1994, 24, 19–41. [Google Scholar] [CrossRef]
- Toppozini, L.; Dies, H.; Deamer, D.W.; Rheinstädter, M.C. Adenosine monophosphate forms ordered arrays in multilamellar lipid matrices: Insights into assembly of nucleic acid for primitive life. PLoS ONE 2013, 8, e62810. [Google Scholar] [CrossRef] [Green Version]
- Himbert, S.; Chapman, M.; Deamer, D.W.; Rheinstädter, M.C. Organization of nucleotides in different environments and the formation of pre-polymers. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Deamer, D. Where Did Life Begin? Testing Ideas in Prebiotic Analogue Conditions. Life 2021, 11, 134. [Google Scholar] [CrossRef]
- Deamer, D.; Damer, B.; Kompanichenko, V. Hydrothermal chemistry and the origin of cellular life. Astrobiology 2019, 19, 1523–1537. [Google Scholar] [CrossRef]
- Deamer, D.W.; Georgiou, C.D. Hydrothermal Conditions and the Origin of Cellular Life. Astrobiology 2015, 15, 1091–1095. [Google Scholar] [CrossRef] [PubMed]
- Deamer, D. Liquid-crystallinenano structures: Organizing matrices for non-enzymatic nucleic acid polymerization. Chem. Soc. Rev. 2012, 41, 5375–5379. [Google Scholar] [CrossRef] [PubMed]
- Hassenkam, T.; Damer, B.; Mednick, G.; Deamer, D. Viroid-sized rings self-assemble from mononucleotides through wet-dry cycling: Implications for the origin of life. bioRxiv 2020, 10, 321. [Google Scholar] [CrossRef] [PubMed]
- Ross, D.S.; Deamer, D. Dry/wet cycling and the thermodynamics and kinetics of prebiotic polymer synthesis. Life 2016, 6, 28. [Google Scholar] [CrossRef] [Green Version]
- Damer, B.; Deamer, D. The hot spring hypothesis for an origin of life. Astrobiology 2020, 20, 429–452. [Google Scholar] [CrossRef] [Green Version]
- Da Silva, L.; Maurel, M.C.; Deamer, D. Salt-promoted synthesis of RNA-like molecules in simulated hydrothermal conditions. J. Mol. Evol. 2015, 80, 86–97. [Google Scholar] [CrossRef]
- Hargrave, M.; Thompson, S.K.; Deamer, D. Computational models of polymer synthesis driven by dehydration/rehydration cycles: Repurination in simulated hydrothermal fields. J. Mol. Evol. 2018, 86, 501–510. [Google Scholar] [CrossRef]
- Deamer, D.W. First Life: Discovering the Connections between Stars, Cells, and How Life Began; University of California Press: Berkeley, CA, USA, 2011. [Google Scholar]
- Damer, B.; Deamer, D. Coupled phases and combinatorial selection in fluctuating hydrothermal pools: A scenario to guide experimental approaches to the origin of cellular life. Life 2015, 5, 872–887. [Google Scholar] [CrossRef] [Green Version]
- DeGuzman, V.; Vercoutere, W.; Shenasa, H.; Deamer, D. Generation of Oligonucleotides Under Hydrothermal Conditions by Non-enzymatic Polymerization. J. Mol. Evol. 2014, 78, 251–262. [Google Scholar] [CrossRef]
- Trinks, H.; Schröder, W.; Biebricher, C.K. Ice and the origin of life. Orig. Life Evol. Biosph. 2005, 35, 429–445. [Google Scholar] [CrossRef] [PubMed]
- Menor-Salván, C.; Marín-Yaseli, M.R. Prebiotic chemistry in eutectic solutions at the water–ice matrix. Chem. Soc. Rev. 2012, 41, 5404–5415. [Google Scholar] [CrossRef] [PubMed]
- Attwater, J.; Wochner, A.; Pinheiro, V.B.; Coulson, A.; Holliger, P. Ice as a protocellular medium for RNA replication. Nat. Commun. 2010, 1, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monnard, P.A.; Kanavarioti, A.; Deamer, D.W. Eutectic phase polymerization of activated ribonucleotide mixtures yields quasi-equimolar incorporation of purine and pyrimidine nucleobases. J. Am. Chem. Soc. 2003, 125, 13734–13740. [Google Scholar] [CrossRef]
- Hazen, R.M. Paleomineralogy of the Hadean Eon: A preliminary species list. Am. J. Sci. 2013, 313, 807–843. [Google Scholar] [CrossRef]
- Bonner, W.A.; Kavasmaneck, P.R.; Martin, F.S.; Flores, J.J. Asymmetric adsorption of alanine by quartz. Science 1974, 186, 143–144. [Google Scholar] [CrossRef]
- Bonner, W.A.; Kavasmaneck, P.R.; Martin, F.S.; Flores, J.J. Asymmetric adsorption by quartz: A model for the prebiotic origin of optical activity. Orig. Life 1975, 6, 367–376. [Google Scholar] [CrossRef]
- Soai, K.; Osanai, S.; Kadowaki, K.; Yonekubo, S.; Shibata, T.; Sato, I. D-and L-quartz-promoted highly enantioselective synthesis of a chiral organic compound. J. Am. Chem. Soc. 1999, 121, 11235–11236. [Google Scholar] [CrossRef]
- Evgenii, K.; Wolfram, T. The role of quartz in the origin of optical activity on Earth. Orig. Life Evol. Biosph. 2000, 30, 431–434. [Google Scholar] [CrossRef]
- Hazen, R.M.; Filley, T.R.; Goodfriend, G.A. Selective adsorption of L-and D-amino acids on calcite: Implications for biochemical homochirality. Proc. Natl. Acad. Sci. USA 2001, 98, 5487–5490. [Google Scholar] [CrossRef]
- Ricardo, A.; Carrigan, M.; Olcott, A.; Benner, S. Borate minerals stabilize ribose. Science 2004, 303, 196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wächtershäuser, G. The case for the chemoautotrophic origin of life in an iron-sulfur world. Orig. Life Evol. Biosph. 1990, 20, 173–176. [Google Scholar] [CrossRef]
- Wächtershäuser, G. The cradle chemistry of life: On the origin of natural products in a pyrite-pulled chemoautotrophic origin of life. Pure Appl. Chem. 1993, 65, 1343–1348. [Google Scholar] [CrossRef]
- Blöchl, E.; Keller, M.; Wächtershäuser, G.; Stetter, K.O. Reactions depending on iron sulfide and linking geochemistry with biochemistry. Proc. Natl. Acad. Sci. USA 1992, 89, 8117–8120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cody, G.D.; Boctor, N.Z.; Filley, T.R.; Hazen, R.M.; Scott, J.H.; Sharma, A.; Yoder, H.S., Jr. Primordial carbonylated iron-sulfur compounds and the synthesis of pyruvate. Science 2000, 289, 1337–1340. [Google Scholar] [CrossRef] [Green Version]
- Summers, D.P.; Chang, S. Prebiotic ammonia from reduction of nitrite by iron (II) on the early Earth. Nature 1993, 365, 630–633. [Google Scholar] [CrossRef] [PubMed]
- Pitsch, S.; Eschenmoser, A.; Gedulin, B.; Hui, S.; Arrhenius, G. Mineral induced formation of sugar phosphates. Orig. Life Evol. Biosph. 1995, 25, 297–334. [Google Scholar] [CrossRef]
- Weber, A.L. Formation of pyrophosphate on hydroxyapatite with thioesters as condensing agents. Biosystems 1982, 15, 183–189. [Google Scholar] [CrossRef]
- Smirnov, A.; Hausner, D.; Laffers, R.; Strongin, D.R.; Schoonen, M.A. Abiotic ammonium formation in the presence of Ni-Fe metals and alloys and its implications for the Hadean nitrogen cycle. Geochem. Trans. 2008, 9, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Pasek, M.A.; Dworkin, J.P.; Lauretta, D.S. A radical pathway for organic phosphorylation during schreibersite corrosion with implications for the origin of life. Geochim. Cosmochim. Acta 2007, 71, 1721–1736. [Google Scholar] [CrossRef]
- Grew, E.S.; Bada, J.L.; Hazen, R.M. Borate minerals and origin of the RNA world. Orig. Life Evol. Biosph. 2011, 41, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Wächtershäuser, G. Pyrite formation, the first energy source for life: A hypothesis. Syst. Appl. Microbiol. 1988, 10, 207–210. [Google Scholar] [CrossRef]
- Wächtershäuser, G. Before enzymes and templates: Theory of surface metabolism. Microbiol. Rev. 1988, 52, 452–484. [Google Scholar] [CrossRef] [PubMed]
- Cairns-Smith, A. Takeover mechanisms and early biochemical evolution. Biosystems 1977, 9, 105–109. [Google Scholar] [CrossRef]
- Cairns-Smith, A.G. Sketches for a mineral genetic material. Elements 2005, 1, 157–161. [Google Scholar] [CrossRef]
- Huber, C.; Wächtershäuser, G. Activated acetic acid by carbon fixation on (Fe, Ni) S under primordial conditions. Science 1997, 276, 245–247. [Google Scholar] [CrossRef] [Green Version]
- Brandes, J.A.; Boctor, N.Z.; Cody, G.D.; Cooper, B.A.; Hazen, R.M.; Yoder, H.S. Abiotic nitrogen reduction on the early Earth. Nature 1998, 395, 365–367. [Google Scholar] [CrossRef]
- Cody, G.; Boctor, N.; Hazen, R.; Brandes, J.; Morowitz, H.J.; Yoder, H., Jr. Geochemical roots of autotrophic carbon fixation: Hydrothermal experiments in the system citric acid, H2O-(±FeS)-(±NiS). Geochim. Cosmochim. Acta 2001, 65, 3557–3576. [Google Scholar] [CrossRef]
- Cody, G.; Boctor, N.; Brandes, J.; Filley, T.; Hazen, R.; Yoder, H., Jr. Assaying the catalytic potential of transition metal sulfides for abiotic carbon fixation. Geochim. Cosmochim. Acta 2004, 68, 2185–2196. [Google Scholar] [CrossRef]
- Huber, C.; Kraus, F.; Hanzlik, M.; Eisenreich, W.; Wächtershäuser, G. Elements of metabolic evolution. Chem. Eur. J. 2012, 18, 2063–2080. [Google Scholar] [CrossRef]
- Cody, G.D. Transition metal sulfides and the origins of metabolism. Annu. Rev. Earth Planet. Sci. 2004, 32, 569. [Google Scholar] [CrossRef]
- Cody, G.D. Geochemical connections to primitive metabolism. Elements 2005, 1, 139–143. [Google Scholar] [CrossRef]
- Berndt, M.E.; Allen, D.E.; Seyfried, W.E., Jr. Reduction of CO2 during serpentinization of olivine at 300 C and 500 bar. Geology 1996, 24, 351–354. [Google Scholar] [CrossRef]
- McCollom, T.M.; Seewald, J.S. A reassessment of the potential for reduction of dissolved CO2 to hydrocarbons during serpentinization of olivine. Geochim. Cosmochim. Acta 2001, 65, 3769–3778. [Google Scholar] [CrossRef]
- Weber, A.L. Prebiotic polymerization: Oxidative polymerization of 2,3-dimercapto-l-propanol on the surface of iron (III) hydroxide oxide. Orig. Life Evol. Biosph. 1995, 25, 53–60. [Google Scholar] [CrossRef]
- Hanczyc, M.M.; Fujikawa, S.M.; Szostak, J.W. Experimental models of primitive cellular compartments: Encapsulation, growth, and division. Science 2003, 302, 618–622. [Google Scholar] [CrossRef] [Green Version]
- Russell, M.J.; Daniel, R.M.; Hall, A.J.; Sherringham, J.A. A hydrothermally precipitated catalytic iron sulphide membrane as a first step toward life. J. Mol. Evol. 1994, 39, 231–243. [Google Scholar] [CrossRef]
- Parsons, I.; Lee, M.R.; Smith, J.V. Biochemical evolution II: Origin of life in tubular microstructures on weathered feldspar surfaces. Proc. Natl. Acad. Sci. USA 1998, 95, 15173–15176. [Google Scholar] [CrossRef] [Green Version]
- Smith, J.V.; Arnold, F.P., Jr.; Parsons, I.; Lee, M.R. Biochemical evolution III: Polymerization on organophilic silica-rich surfaces, crystal-chemical modeling, formation of first cells, and geological clues. Proc. Natl. Acad. Sci. USA 1999, 96, 3479–3485. [Google Scholar] [CrossRef] [Green Version]
- Lahav, N.; White, D.; Chang, S. Peptide formation in the prebiotic era: Thermal condensation of glycine in fluctuating clay environments. Science 1978, 201, 67–69. [Google Scholar] [CrossRef]
- Huber, C.; Wächtershäuser, G. Peptides by activation of amino acids with CO on (Ni, Fe) S surfaces: Implications for the origin of life. Science 1998, 281, 670–672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huber, C.; Eisenreich, W.; Hecht, S.; Wächtershäuser, G. A possible primordial peptide cycle. Science 2003, 301, 938–940. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.V. Biochemical evolution I: Polymerization on internal, organophilic silica surfaces of dealuminated zeolites and feldspars. Proc. Natl. Acad. Sci. USA 1998, 95, 3370–3375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jonsson, C.M.; Jonsson, C.L.; Sverjensky, D.A.; Cleaves, H.J.; Hazen, R.M. Attachment of l-glutamate to rutile (α-TiO2): A potentiometric, adsorption, and surface complexation study. Langmuir 2009, 25, 12127–12135. [Google Scholar] [CrossRef] [PubMed]
- Ferris, J.P. Mineral catalysis and prebiotic synthesis: Montmorillonite-catalyzed formation of RNA. Elements 2005, 1, 145–149. [Google Scholar] [CrossRef]
- Ferris, J.P. Montmorillonite-catalysed formation of RNA oligomers: The possible role of catalysis in the origins of life. Philos. Trans. R. Soc. Biol. Sci. 2006, 361, 1777–1786. [Google Scholar] [CrossRef] [Green Version]
- Ferris, J.P.; Ertem, G. Oligomerization of ribonucleotides on montmorillonite: Reaction of the 5′-phosphorimidazolide of adenosine. Science 1992, 257, 1387–1389. [Google Scholar] [CrossRef]
- Ferris, J.P. Catalysis and prebiotic RNA synthesis. Orig. Life Evol. Biosph. 1993, 23, 307–315. [Google Scholar] [CrossRef]
- Ferris, J.P.; Ertem, G. Montmorillonite catalysis of RNA oligomer formation in aqueous solution. A model for the prebiotic formation of RNA. J. Am. Chem. Soc. 1993, 115, 12270–12275. [Google Scholar] [CrossRef]
- Ertem, G.; Ferris, J.P. Synthesis of RNA oligomers on heterogeneous templates. Nature 1996, 379, 238–240. [Google Scholar] [CrossRef]
- Ertem, G.; Ferris, J.P. Template-directed synthesis using the heterogeneous templates produced by montmorillonite catalysis. A possible bridge between the prebiotic and RNA worlds. J. Am. Chem. Soc. 1997, 119, 7197–7201. [Google Scholar] [CrossRef] [PubMed]
- Holm, N.G.; Ertem, G.; Ferris, J.P. The binding and reactions of nucleotides and polynucleotides on iron oxide hydroxide polymorphs. Orig. Life Evol. Biosph. 1993, 23, 195–215. [Google Scholar] [CrossRef] [PubMed]
- Acevedo, O.L.; Orgel, L.E. Template-directed oligonucleotide ligation on hydroxylapatite. Nature 1986, 321, 790–792. [Google Scholar] [CrossRef]
- Ciciriello, F.; Costanzo, G.; Crestini, C.; Saladino, R.; Di Mauro, E. Origin of informational polymers and the search for non-terran life: Protection of the polymeric state of DNA by phosphate minerals. Astrobiology 2007, 7, 616–630. [Google Scholar] [CrossRef]
- Benner, S.A.; Hutter, D. Phosphates, DNA, and the search for nonterrean life: A second generation model for genetic molecules. Bioorg. Chem. 2002, 30, 62–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, W.; Ferris, J.P. One-step, regioselective synthesis of up to 50-mers of RNA oligomers by montmorillonite catalysis. J. Am. Chem. Soc. 2006, 128, 8914–8919. [Google Scholar] [CrossRef]
- Gilbert, W. Origin of life: The RNA world. Nature 1986, 319, 618. [Google Scholar] [CrossRef]
- Rich, A. On the problems of evolution and biochemical information transfer. Horizons Biochem. 1962, 103–126. [Google Scholar]
- Cech, T.R. A model for the RNA-catalyzed replication of RNA. Proc. Natl. Acad. Sci. USA 1986, 83, 4360–4363. [Google Scholar] [CrossRef] [Green Version]
- Cech, T.R. The RNA worlds in context. Cold Spring Harb. Perspect. Biol. 2012, 4, a006742. [Google Scholar] [CrossRef] [Green Version]
- Hud, N.V.; Cafferty, B.J.; Krishnamurthy, R.; Williams, L.D. The origin of RNA and “my grandfather’s axe”. Chem. Biol. 2013, 20, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Joyce, G.F. Forty years of in vitro evolution. Angew. Chem. Int. Ed. 2007, 46, 6420–6436. [Google Scholar] [CrossRef]
- Biebricher, C.K.; Orgel, L.E. An RNA that multiplies indefinitely with DNA-dependent RNA polymerase: Selection from a random copolymer. Proc. Natl. Acad. Sci. USA 1973, 70, 934–938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biebricher, C.K.; Eigen, M.; Gardiner, W.C., Jr. Kinetics of RNA replication: Competition and selection among self-replicating RNA species. Biochemistry 1985, 24, 6550–6560. [Google Scholar] [CrossRef] [PubMed]
- Robertson, D.L.; Joyce, G.F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 1990, 344, 467–468. [Google Scholar] [CrossRef]
- Beaudry, A.A.; Joyce, G.F. Directed evolution of an RNA enzyme. Science 1992, 257, 635–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Neveu, M.; Kim, H.J.; Benner, S.A. The “strong” RNA world hypothesis: Fifty years old. Astrobiology 2013, 13, 391–403. [Google Scholar] [CrossRef] [Green Version]
- Ferris, J.P.; Ertem, G.; Agarwal, V.K. The adsorption of nucleotides and polynucleotides on montmorillonite clay. Orig. Life Evol. Biosph. 1989, 19, 153–164. [Google Scholar] [CrossRef]
- Franchi, M.; Ferris, J.P.; Gallori, E. Cations as mediators of the adsorption of nucleic acids on clay surfaces in prebiotic environments. Orig. Life Evol. Biosph. 2003, 33, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Pedreira-Segade, U.; Feuillie, C.; Pelletier, M.; Michot, L.J.; Daniel, I. Adsorption of nucleotides onto ferromagnesian phyllosilicates: Significance for the origin of life. Geochim. Cosmochim. Acta 2016, 176, 81–95. [Google Scholar] [CrossRef]
- Mast, C.B.; Schink, S.; Gerland, U.; Braun, D. Escalation of polymerization in a thermal gradient. Proc. Natl. Acad. Sci. USA 2013, 110, 8030–8035. [Google Scholar] [CrossRef] [Green Version]
- Klein, C.; Dutrow, B.; Dana, J.D. The 23rd Edition of the Manual of Mineral Science: (After James D. Dana); Number 549 KLE; John Wiley: Hoboken, NJ, USA, 2007. [Google Scholar]
- Scappini, F.; Casadei, F.; Zamboni, R.; Franchi, M.; Gallori, E.; Monti, S. Protective effect of clay minerals on adsorbed nucleic acid against UV radiation: Possible role in the origin of life. Int. J. Astrobiol. 2004, 3, 17–19. [Google Scholar] [CrossRef]
- Kasprzhitskii, A.; Lazorenko, G.; Kharytonau, D.S.; Osipenko, M.A.; Kasach, A.A.; Kurilo, I.I. Adsorption mechanism of aliphatic amino acids on kaolinite surfaces. Appl. Clay Sci. 2022, 226, 106566. [Google Scholar] [CrossRef]
- Strigunkova, T.; Lavrentiev, G.; Otroshchenko, V. Abiogenic synthesis of oligonucleotides on kaolinite under the action of ultraviolet radiation. J. Mol. Evol. 1986, 23, 290–293. [Google Scholar] [CrossRef]
- Zhang, C.; Yu, Z.G.; Zeng, G.M.; Jiang, M.; Yang, Z.Z.; Cui, F.; Zhu, M.Y.; Shen, L.Q.; Hu, L. Effects of sediment geochemical properties on heavy metal bioavailability. Environ. Int. 2014, 73, 270–281. [Google Scholar] [CrossRef]
- Murray, H.H. Overview—Clay mineral applications. Appl. Clay Sci. 1991, 5, 379–395. [Google Scholar] [CrossRef]
- Nesse, W.D. Introduction to Mineralogy; Oxford University Press: Oxford, UK, 2012. [Google Scholar]
- Hansma, H.G. The power of crowding for the origins of life. Orig. Life Evol. Biosph. 2014, 44, 307–311. [Google Scholar] [CrossRef]
- Hansma, H.G. Possible origin of life between mica sheets: Does life imitate mica? J. Biomol. Struct. Dyn. 2013, 31, 888–895. [Google Scholar] [CrossRef] [Green Version]
- Hansma, H.G. Possible origin of life between mica sheets. J. Theor. Biol. 2010, 266, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Hazen, R.M.; Papineau, D.; Bleeker, W.; Downs, R.T.; Ferry, J.M.; McCoy, T.J.; Sverjensky, D.A.; Yang, H. Mineral evolution. Am. Mineral. 2008, 93, 1693–1720. [Google Scholar] [CrossRef]
- Cleaves, H.J., II; Scott, A.M.; Hill, F.C.; Leszczynski, J.; Sahai, N.; Hazen, R. Mineral–organic interfacial processes: Potential roles in the origins of life. Chem. Soc. Rev. 2012, 41, 5502–5525. [Google Scholar] [CrossRef]
- Dalai, P.; Sahai, N. Mineral–lipid interactions in the origins of life. Trends Biochem. Sci. 2019, 44, 331–341. [Google Scholar] [CrossRef]
- Nittler, L.R. Presolar stardust in meteorites: Recent advances and scientific frontiers. Earth Planet. Sci. Lett. 2003, 209, 259–273. [Google Scholar] [CrossRef]
- Cuéllar-Cruz, M.; Moreno, A. Synthesis of Crystalline Silica–Carbonate Biomorphs of Ba (II) under the Presence of RNA and Positively and Negatively Charged ITO Electrodes: Obtainment of Graphite via Bioreduction of CO2 and Its Implications to the Chemical Origin of Life on Primitive Earth. ACS Omega 2020, 5, 5460–5469. [Google Scholar] [PubMed] [Green Version]
- Sowerby, S.J.; Cohn, C.A.; Heckl, W.M.; Holm, N.G. Differential adsorption of nucleic acid bases: Relevance to the origin of life. Proc. Natl. Acad. Sci. USA 2001, 98, 820–822. [Google Scholar] [CrossRef] [Green Version]
- Ortmann, F.; Schmidt, W.G.; Bechstedt, F. Attracted by long-range electron correlation: Adenine on graphite. Phys. Rev. Lett. 2005, 95, 186101. [Google Scholar] [CrossRef] [Green Version]
- BIOVIA, Dassault Systemes. BIOVIA Discovery Studio Visualizer; Dassault Systèmes: San Diego, CA, USA, 2021. [Google Scholar]
- Gasteiger, J.; Marsili, M. Iterative partial equalization of orbital electronegativity—A rapid access to atomic charges. Tetrahedron 1980, 36, 3219–3228. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; et al. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 2016, 12, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.R.; Brooks, C.L., III; Mackerell, A.D., Jr.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Šolc, R.; Gerzabek, M.H.; Lischka, H.; Tunega, D. Wettability of kaolinite (001) surfaces—Molecular dynamic study. Geoderma 2011, 169, 47–54. [Google Scholar] [CrossRef]
- Hench, L.L.; West, J.K. Inorganic pathways for biosynthesis: A molecular orbital modeling approach. J. Vac. Sci. Technol. 1994, 12, 2962–2965. [Google Scholar] [CrossRef]
- Gambino, G.L.; Lombardo, G.M.; Grassi, A.; Marletta, G. Molecular modeling of interactions between L-lysine and a hydroxylated quartz surface. J. Phys. Chem. B 2004, 108, 2600–2607. [Google Scholar] [CrossRef]
- Gambino, G.L.; Grassi, A.; Marletta, G. Molecular modeling of interactions between L-lysine and functionalized quartz surfaces. J. Phys. Chem. B 2006, 110, 4836–4845. [Google Scholar] [CrossRef]
- Lomenech, C.; Bery, G.; Costa, D.; Stievano, L.; Lambert, J.F. Theoretical and experimental study of the adsorption of neutral glycine on silica from the gas phase. ChemPhysChem 2005, 6, 1061–1070. [Google Scholar] [CrossRef] [Green Version]
- Rimola, A.; Tosoni, S.; Sodupe, M.; Ugliengo, P. Peptide bond formation activated by the interplay of Lewis and Brønsted catalysts. Chem. Phys. Lett. 2005, 408, 295–301. [Google Scholar] [CrossRef]
- Rimola, A.; Tosoni, S.; Sodupe, M.; Ugliengo, P. Does Silica Surface Catalyse Peptide Bond Formation? New Insights from First-Principles Calculations. ChemPhysChem 2006, 7, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Rimola, A.; Sodupe, M.; Ugliengo, P. Aluminosilicate surfaces as promoters for peptide bond formation: An assessment of Bernal’s hypothesis by ab Initio methods. J. Am. Chem. Soc. 2007, 129, 8333–8344. [Google Scholar] [CrossRef] [PubMed]
- Costa, D.; Lomenech, C.; Meng, M.; Stievano, L.; Lambert, J.F. Microsolvation of glycine by silanol ligands: A DFT study. J. Mol. Struct. Theochem 2007, 806, 253–259. [Google Scholar] [CrossRef]
- Stievano, L.; Piao, L.Y.; Lopes, I.; Meng, M.; Costa, D.; Lambert, J.F. Glycine and lysine adsorption and reactivity on the surface of amorphous silica. Eur. J. Mineral. 2007, 19, 321–331. [Google Scholar] [CrossRef]
- Collins, J.R.; Loew, G.H.; Luke, B.T.; White, D.H. Theoretical investigation of the role of clay edges in prebiotic peptide bond formation. Orig. Life Evol. Biosph. 1988, 18, 107–119. [Google Scholar] [CrossRef]
- Yu, C.H.; Newton, S.Q.; Miller, D.M.; Teppen, B.J.; Schäfer, L. Ab initio study of the nonequivalence of adsorption of D-and L-peptides on clay mineral surfaces. Struct. Chem. 2001, 12, 393–398. [Google Scholar] [CrossRef]
- Pérez-Villa, A.; Saitta, A.M.; Georgelin, T.; Lambert, J.F.; Guyot, F.; Maurel, M.C.; Pietrucci, F. Synthesis of RNA nucleotides in plausible prebiotic conditions from ab initio computer simulations. J. Phys. Chem. Lett. 2018, 9, 4981–4987. [Google Scholar] [CrossRef]
- Bertram, L.; Roberts, S.J.; Powner, M.W.; Szabla, R. Photochemistry of 2-thiooxazole: A plausible prebiotic precursor to RNA nucleotides. Phys. Chem. Chem. Phys. 2022, 24, 21406–21416. [Google Scholar] [CrossRef]
- Niether, D.; Afanasenkau, D.; Dhont, J.K.; Wiegand, S. Accumulation of formamide in hydrothermal pores to form prebiotic nucleobases. Proc. Natl. Acad. Sci. USA 2016, 113, 4272–4277. [Google Scholar] [CrossRef] [Green Version]
- Ferris, J.P.; Hill, A.R.; Liu, R.; Orgel, L.E. Synthesis of long prebiotic oligomers on mineral surfaces. Nature 1996, 381, 59–61. [Google Scholar] [CrossRef] [Green Version]
- Jerome, C.A.; Kim, H.J.; Mojzsis, S.J.; Benner, S.A.; Biondi, E. Catalytic Synthesis of Polyribonucleic Acid on Prebiotic Rock Glasses. Astrobiology 2022, 22. [Google Scholar] [CrossRef] [PubMed]
- Biondi, E.; Furukawa, Y.; Kawai, J.; Benner, S.A. Adsorption of RNA on mineral surfaces and mineral precipitates. Beilstein J. Org. Chem. 2017, 13, 393–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biondi, E.; Howell, L.; Benner, S.A. Opal adsorbs and stabilizes RNA–a hierarchy of prebiotic silica minerals. Synlett 2017, 28, 84–88. [Google Scholar] [CrossRef] [Green Version]
- Biondi, E.; Branciamore, S.; Maurel, M.C.; Gallori, E. Montmorillonite protection of an UV-irradiated hairpin ribozyme: Evolution of the RNA world in a mineral environment. BMC Evol. Biol. 2007, 7, 1–7. [Google Scholar] [CrossRef]
- Sowerby, S.J.; Petersen, G.B. Scanning tunnelling microscopy and molecular modelling of xanthine monolayers self-assembled at the solid-liquid interface: Relevance to the origin of life. Orig. Life Evol. Biosph. 1999, 29, 597–614. [Google Scholar] [CrossRef]
- Holterman, H. Kinetics and Evaporation of Water Drops in Air; Number 2003-12; IMAG: Wageningen, The Netherlands, 2003. [Google Scholar]
- Ferris, J.; Joshi, P.; Wang, K.J.; Miyakawa, S.; Huang, W. Catalysis in prebiotic chemistry: Application to the synthesis of RNA oligomers. Adv. Space Res. 2004, 33, 100–105. [Google Scholar] [CrossRef]
- Kloprogge, J. Raman spectroscopy of clay minerals. In Developments in Clay Science; Elsevier: Amsterdam, The Netherlands, 2017; Volume 8, pp. 150–199. [Google Scholar]
- Hess, B. P-LINCS: A parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 2008, 4, 116–122. [Google Scholar] [CrossRef]
- Nosé, S. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 1984, 52, 255–268. [Google Scholar] [CrossRef]
- Nosé, S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984, 81, 511–519. [Google Scholar] [CrossRef] [Green Version]
- Hoover, W.G. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A 1985, 31, 1695. [Google Scholar] [CrossRef] [Green Version]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]













| Material | Mineral | Formula |
|---|---|---|
| Clay | Montmorillonite | (K, Na)[SiO][AlMgO(OH)] |
| Kaolinite | AlSiO(OH) | |
| Pyrophyllite | AlSiO(OH)) | |
| Mica | Muscovite | (KAl(AlSi)O(OH)) |
| Phosphate minerals | Hydroxyapatite | Ca(PO)(OH) |
| Silica | -quartz | SiO |
| Metal oxides | Corundum | AlO |
| Periclase | MgO | |
| Carbonaceous material | Graphite | C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dujardin, A.; Himbert, S.; Pudritz, R.; Rheinstädter, M.C. The Formation of RNA Pre-Polymers in the Presence of Different Prebiotic Mineral Surfaces Studied by Molecular Dynamics Simulations. Life 2023, 13, 112. https://doi.org/10.3390/life13010112
Dujardin A, Himbert S, Pudritz R, Rheinstädter MC. The Formation of RNA Pre-Polymers in the Presence of Different Prebiotic Mineral Surfaces Studied by Molecular Dynamics Simulations. Life. 2023; 13(1):112. https://doi.org/10.3390/life13010112
Chicago/Turabian StyleDujardin, Alix, Sebastian Himbert, Ralph Pudritz, and Maikel C. Rheinstädter. 2023. "The Formation of RNA Pre-Polymers in the Presence of Different Prebiotic Mineral Surfaces Studied by Molecular Dynamics Simulations" Life 13, no. 1: 112. https://doi.org/10.3390/life13010112
APA StyleDujardin, A., Himbert, S., Pudritz, R., & Rheinstädter, M. C. (2023). The Formation of RNA Pre-Polymers in the Presence of Different Prebiotic Mineral Surfaces Studied by Molecular Dynamics Simulations. Life, 13(1), 112. https://doi.org/10.3390/life13010112