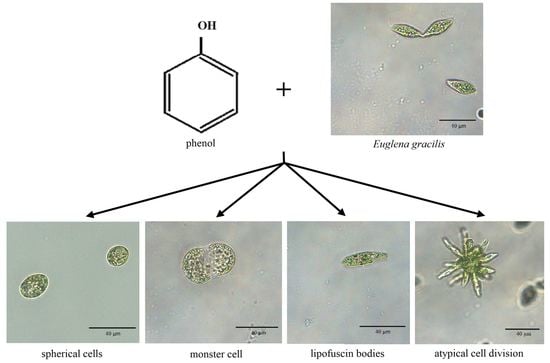
The Influence of Phenol on the Growth, Morphology and Cell Division of Euglena gracilis
Abstract
:1. Introduction
2. Material and Methods
2.1. Cultivation Conditions, Phenol Treatment and Monitored Parameters
2.2. Light Microscopy
2.3. Transmission Electron Microscopy (TEM)
2.4. Confocal Microscopy of Cells with Atypical Cell Division
2.5. Statistical Analysis
3. Results
3.1. The Effect of Phenol on E. gracilis Cell Growth
3.2. The Effect of Phenol on the Morphology of E. gracilis
3.3. Phenol Induces Atypical Cell Division of E. gracilis
3.4. Statistical Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vesteg, M.; Hadariová, L.; Horváth, A.; Estraňo, C.E.; Schwartzbach, S.D.; Krajčovič, J. Comparative molecular cell biology of phototrophic euglenids and parasitic trypanosomatids sheds light on the ancestor of Euglenozoa. Biol. Rev. Camb. Philos. Soc. 2019, 94, 1701–1721. [Google Scholar] [CrossRef] [PubMed]
- Adl, S.M.; Bass, D.; Lane, C.E.; Lukeš, J.; Schoch, C.L.; Smirnov, A.; Agatha, S.; Berney, C.; Brown, M.W.; Burki, F.; et al. Revisions to the classification, nomenclature and diversity of eukaryotes. J. Eukaryot. Microbiol. 2019, 66, 4–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turmel, M.; Gagnon, M.C.; O’Kelly, C.J.; Otis, C.; Lemieux, C. The chloroplast genomes of the green algae Pyramimonas, Monomastix, and Pycnococcus shed new light on the evolutionary history of prasinophytes and the origin of the secondary chloroplasts of euglenids. Mol. Biol. Evol. 2009, 26, 631–648. [Google Scholar] [CrossRef] [Green Version]
- Vesteg, M.; Vacula, R.; Steiner, J.M.; Mateášiková, B.; Löffelhardt, W.; Brejová, B.; Krajčovič, J. A possible role for short introns in the acquisition of stroma-targeting peptides in the flagellate Euglena gracilis. DNA Res. 2010, 17, 223–231. [Google Scholar] [CrossRef] [Green Version]
- Vesteg, M.; Vacula, R.; Burey, S.; Löffelhardt, W.; Drahovská, H.; Martin, W.; Krajčovič, J. Expression of nucleus-encoded genes for chloroplast proteins in the flagellate Euglena gracilis. J. Eukaryot. Microbiol. 2009, 56, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Mateášiková-Kováčová, B.; Vesteg, M.; Drahovská, H.; Záhonová, K.; Vacula, R.; Krajčovič, J. Nucleus-encoded mRNAs for chloroplast proteins GapA, PetA a PsbO are trans-spliced in the flagellate Euglena gracilis irrespective of light and plastid function. J. Eukaryot. Microbiol. 2012, 59, 651–653. [Google Scholar] [CrossRef]
- Krnáčová, K.; Vinarčíková, M.; Rýdlová, I.; Krajčovič, J.; Vesteg, M.; Horváth, A. Characterization of oxidative phosphorylation enzymes in Euglena gracilis and its white mutant strain WgmZOflL. FEBS Lett. 2015, 589, 687–694. [Google Scholar] [CrossRef] [Green Version]
- Richter, P.; Börnig, A.; Streb, C.; Ntefidou, M.; Lebert, M.; Häder, D.P. Effects of increased salinity on gravitaxis in Euglena gracilis. J. Plant Physiol. 2003, 160, 651–656. [Google Scholar] [CrossRef]
- Hayashi, H.; Narumi, I.; Wada, S.; Kikuchi, M.; Furuta, M.; Uehara, K.; Watanabe, H. Light dependency of resistance to ionizing radiation in Euglena gracilis. J. Plant Physiol. 2004, 161, 1101–1106. [Google Scholar] [CrossRef]
- Krajčovič, J.; Vesteg, M.; Schwartzbach, S.D. Euglenoid flagellates: A multifaceted biotechnology platform. J. Biotechnol. 2015, 202, 135–145. [Google Scholar] [CrossRef]
- Gissibl, A.; Sun, A.; Care, A.; Nevalainen, H.; Sunna, A. Bioproducts from Euglena gracilis: Synthesis and applications. Front. Bioeng. Biotechnol. 2019, 7, 108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khatiwada, B.; Sunna, A.; Nevalainen, H. Molecular tools and applications of Euglena gracilis: From biorefineries to bioremediation. Biotechnol. Bioeng. 2020, 117, 3952–3967. [Google Scholar] [CrossRef] [PubMed]
- Lukáčová, A.; Beck, T.; Koptašiková, L.; Benda, A.; Tomečková, L.; Trniková, M.; Lihanová, D.; Steiner, J.M.; Krajčovič, J.; Vesteg, M. Euglena gracilis can grow in the mixed culture containing Cladosporium westerdijkiae, Lysinibacillus boronitolerans and Pseudobacillus badius without the addition of vitamins B1 and B12. J. Biotechnol. 2022, 351, 50–59. [Google Scholar] [CrossRef]
- Lihanová, D.; Lukáčová, A.; Beck, T.; Jedlička, A.; Vešelényiová, D.; Krajčovič, J.; Vesteg, M. Versatile biotechnological applications of Euglena gracilis. World J. Microbiol. Biotechnol. 2023, 39, 133. [Google Scholar] [CrossRef]
- Stallwitz, E.; Häder, D.P. Motility and phototactic orientation of the flagellate Euglena gracilis impaired by heavy-metal ions. J. Photochem. Photobiol. B Biol. 1993, 18, 67–74. [Google Scholar] [CrossRef]
- Rocchetta, I.; Küpper, H. Chromium- and copper-induced inhibition of photosynthesis in Euglena gracilis analysed on the single-cell level by fluorescence kinetic microscopy. New Phytol. 2009, 182, 405–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-García, J.D.; Peña-Sanabria, K.A.; Sánchez-Thomas, R.; Moreno-Sánchez, R. Nickel accumulation by the green algae-like Euglena gracilis. J. Hazard. Mater. 2018, 343, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Arthur, D.M.; Sichani, H.T.; Xia, Q.; Ng, J.C. Assessing benzene-induced toxicity on wild type Euglena gracilis Z and its mutant strain SMZ. Chemosphere 2013, 93, 2381–2389. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Lee, J.W.; Sichani, H.T.; Ng, J.C. Toxic effects of individual and combined effects of BTEX on Euglena gracilis. J. Hazard. Mater. 2015, 284, 10–18. [Google Scholar] [CrossRef]
- Krajčovič, J.; Ebringer, L.; Schwartzbach, S.D. Reversion of endosymbiosis? In Symbiosis: Mechanisms and Models. Cellular Origin in Extreme Habitats; Seckbach, J., Ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2002; Volume 4, pp. 185–206. [Google Scholar]
- Hadariová, L.; Vesteg, M.; Birčák, E.; Schwartzbach, S.D.; Krajčovič, J. An intact plastid genome is essential for the survival of colorless Euglena longa but not Euglena gracilis. Curr. Genet. 2017, 63, 331–341. [Google Scholar] [CrossRef]
- Hadariová, L.; Vesteg, M.; Hampl, V.; Krajčovič, J. Reductive evolution of chloroplasts in non-photosynthetic plants, algae and protists. Curr. Genet. 2018, 64, 365–387. [Google Scholar] [CrossRef]
- Patková, J.; Vojtíšek, M.; Tůma, J.; Vožeh, F.; Knotková, J.; Santorová, P.; Wilhelm, J. Evaluation of lipofuscin-like pigments as an index of lead-induced oxidative damage in the brain. Exp. Toxicol. Pathol. 2012, 64, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Vaschenko, M.A.; Zhadan, P.M.; Aminin, D.L.; Almyashova, T.N. Lipofuscin-like pigment in gonads of sea urchin Strongylocentrotus intermedius as a potential biomarker of marine pollution: A field study. Arch. Environ. Contam. Toxicol. 2012, 62, 599–613. [Google Scholar] [CrossRef]
- Sun, R.; Wang, Y.; Ni, Y.; Kokot, S. Spectrophotometric analysis of phenols, which involves a hemin–graphene hybrid nanoparticles with peroxidase-like activity. J. Hazard. Mater. 2014, 266, 60–67. [Google Scholar] [CrossRef]
- Mohd, A. Presence of phenol in wastewater effluent and its removal: An overview. Int. J. Environ. Chem. 2020, 102, 1362–1384. [Google Scholar] [CrossRef]
- García García, I.; Bonilla Venceslada, J.L.; Jiménez Peña, P.R.; Ramos Gómez, E. Biodegradation of phenol compounds in vinasse using Aspergillus terreus and Geotrichum candidum. Water Res. 1997, 31, 2005–2011. [Google Scholar] [CrossRef]
- Jusoh, N.; Razali, F. Microbial consortia from residential wastewater for bioremediation of phenol in a chemostat. J. Teknol. 2008, 4, 51–60. [Google Scholar]
- Mohammadi, S.; Kargari, A.; Sanaeepur, H.; Abbassian, K.; Najafi, A.; Mofarrah, E. Phenol removal from industrial wastewaters: A short review. Desalin. Water Treat. 2015, 53, 2215–2234. [Google Scholar] [CrossRef]
- Surkatti, R.; Al-Zuhair, S. Microalgae cultivation for phenolic compounds removal. Environ. Sci. Pollut. Res. 2018, 25, 33936–33956. [Google Scholar] [CrossRef]
- Anku, W.W.; Mamo, M.A.; Govender, P.P. Phenolic compounds in water: Sources, reactivity, toxicity and treatment methods. In Phenolic Compounds—Natural Sources, Importance and Applications; Soto-Hernández, M., Palma-Tenango, M., Garcia-Mateos, M.d.R., Eds.; InTech: London, UK, 2017; pp. 419–443. [Google Scholar] [CrossRef] [Green Version]
- Cramer, M.; Myers, J. Growth and photosynthetic characteristics of Euglena gracilis. Arch. Microbiol. 1952, 17, 384–403. [Google Scholar] [CrossRef]
- Buetow, D.E.; Padilla, G.M. Growth of Astasia longa on ethanol. I. Effects of ethanol on generation time, population density and biochemical profile. J. Protozool. 1963, 10, 121–123. [Google Scholar] [CrossRef] [PubMed]
- Latt, S.A.; Stetten, G.; Juergens, L.A.; Willard, H.F.; Scher, C.D. Recent developments in the detection of deoxyribonucleic acid synthesis by 33258 Hoechst fluorescence. J. Histochem. Cytochem. 1975, 23, 493–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kottuparambil, S.; Kim, Y.J.; Choi, H.; Kim, M.S.; Park, A.; Park, J.; Shin, W.; Han, T. A rapid phenol toxicity test based on photosynthesis and movement of the freshwater flagellate, Euglena agilis Carter. Aquat. Toxicol. 2014, 155, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, S.J.; Kaware, J.P. Review on research for removal of phenol from wastewater. Int. J. Sci. Res. Publ. 2013, 3, 1–5. [Google Scholar]
- Wei, X.; Gilevska, T.; Wetzig, F.; Dorer, C.; Richnow, H.H.; Vogt, C. Characterization of phenol and cresol biodegradation by compound-specific stable isotope analysis. Environ. Pollut. 2016, 210, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Suzuki, T. Cadmium-induced abnormality in strains of Euglena gracilis: Morphological alteration and its prevention by zinc and cyanocobalamin. Comp. Biochem. Physiol. C 2001, 130, 29–39. [Google Scholar] [CrossRef]
- Terman, A.; Brunk, U.T. Lipofuscin: Mechanisms of formation and increase with age. APMIS 1998, 106, 265–276. [Google Scholar] [CrossRef]
- Pilátová, J.; Mojzeš, P.; Hurková, K.; Schwarzerová, K. Enigmatic organelles of cellular senescence—The great synthesis. In Proceedings of the 51st Jírovec’s Protozoological Days, Hotel Svratka, Czech Republic, 20–24 June 2022; p. 27. Available online: https://drive.google.com/file/d/1pkzIshqR05--e-xLP5MR1H0ICWbsacsb/view (accessed on 27 July 2023).
- Eliáš, M.; Amaral, R.; Fawley, K.P.; Fawley, M.W.; Němcová, Y.; Neustupa, J.; Přibyl, P.; Santos, L.M.A.; Ševčíková, T. Eustigmatophyceae. In Handbook of the Protists; Archibald, J., Simpson, A., Slamovits, C., Eds.; Springer: Cham, Switzerland, 2017; pp. 367–406. [Google Scholar] [CrossRef]
- Zhou, J.; Fritz, L. The PAS/accumulation bodies in Prorocentrum lima and Prorocentrum maculosum (Dinophyceae) are dinoflagellate lysosomes. J. Phycol. 1994, 30, 39–44. [Google Scholar] [CrossRef]
- Vesteg, M.; Krajčovič, J. The falsifiability of the models for the origin of eukaryotes. Curr. Genet. 2011, 57, 367–390. [Google Scholar] [CrossRef]
- Vesteg, M.; Šándorová, Z.; Krajčovič, J. Selective forces for the origin of spliceosomes. J. Mol. Evol. 2012, 74, 226–231. [Google Scholar] [CrossRef]
- Leedale, G.F. The nucleus in Euglena. In The Biology of Euglena 1. General Biology and Ultrastructure; Buetow, D.E., Ed.; Academic Press: New York, NY, USA, 1968; pp. 185–242. [Google Scholar]
- Vannini, G.L.; Fasulo, M.P.; Bruni, A. First observations on the cytological changes induced in Euglena gracilis by coumarin treatment. Z. Pflanzenphysiol. 1977, 84, 183–187. [Google Scholar] [CrossRef]
- Fasulo, M.P.; Bassi, M.; Donini, A. Cytotoxic effects of hexavalent chromium in Euglena gracilis. II. Biological and ultrastructural studies. Protoplasma 1982, 114, 35–43. [Google Scholar] [CrossRef]
- Zakryś, B. Aberrations in development of strain, Z’, of Euglena gracilis. Bull. Acad. Sci. Polon. Ser. Sci. Biol. 1980, 28, 117–120. [Google Scholar]
- Zakryś, B. Aberrative divisions of Euglena and their phylogenetic implications. Nova Hedw. 1983, 38, 471–476. [Google Scholar]
- Zakryś, B. The nuclear behavior during abnormal cell division in Euglena viridis Ehrbg. Nova Hedw. 1986, 42, 591–596. [Google Scholar]










Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lukáčová, A.; Lihanová, D.; Beck, T.; Alberty, R.; Vešelényiová, D.; Krajčovič, J.; Vesteg, M. The Influence of Phenol on the Growth, Morphology and Cell Division of Euglena gracilis. Life 2023, 13, 1734. https://doi.org/10.3390/life13081734
Lukáčová A, Lihanová D, Beck T, Alberty R, Vešelényiová D, Krajčovič J, Vesteg M. The Influence of Phenol on the Growth, Morphology and Cell Division of Euglena gracilis. Life. 2023; 13(8):1734. https://doi.org/10.3390/life13081734
Chicago/Turabian StyleLukáčová, Alexandra, Diana Lihanová, Terézia Beck, Roman Alberty, Dominika Vešelényiová, Juraj Krajčovič, and Matej Vesteg. 2023. "The Influence of Phenol on the Growth, Morphology and Cell Division of Euglena gracilis" Life 13, no. 8: 1734. https://doi.org/10.3390/life13081734
APA StyleLukáčová, A., Lihanová, D., Beck, T., Alberty, R., Vešelényiová, D., Krajčovič, J., & Vesteg, M. (2023). The Influence of Phenol on the Growth, Morphology and Cell Division of Euglena gracilis. Life, 13(8), 1734. https://doi.org/10.3390/life13081734