Radiomics Diagnostic Tool Based on Deep Learning for Colposcopy Image Classification
Abstract
:1. Introduction
2. Related Work
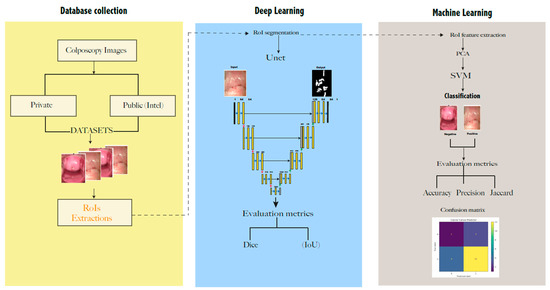
3. Materials and Methods
3.1. Datasets
- (i)
- The public database “Intel & Mobile ODT Cervical Cancer Screening” from Kaggle community developers https://www.kaggle.com/competitions/intel-mobileodt-cervical-cancer-screening/data (accessed on 1 October 2021). It contains 1481 cervix images divided into two categories based on their visual aspect: normal (specified as “considered non-cancerous”) and abnormal. From these, 460 were taken as a training set in this work.
- (ii)
- A private dataset was collected from women from a rural community in Ecuador. All of the images were anonymized and collected from June to December 2020, as a part of the project CAMIE—https://www.camieproject.com/ (accessed on 2 June 2022). Table 1 shows the classification of two image datasets according to their negative or positive diagnosis of lesions.
3.2. Private Data Collection
3.3. Data Augmentation
3.4. Presegmentation: Cropping the Regions of Interest (RoI)
3.5. Segmentation
3.6. Features of the Extraction and Classification
4. Results
4.1. Cervix Image Segmentation with Unet
4.2. Cervix Image Classification with SVM
4.3. Graphical Interface
4.4. Visual Comparison
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AUC | area under the curve |
| CC | cervical cancer |
| CNN | convolutional neural network |
| CAMIE | Cáncer Auto Muestreo Igualdad Empoderamiento |
| CIN | cervical intraepithelial neoplasms |
| CAD | computer-aided diagnosis/detection |
| DL | deep learning |
| FCN | full connected layer |
| GAN | generative adversarial network |
| HPV | human papillomavirus |
| IoU | intersection over union |
| PPV | positive predictive value |
| PCA | principal component analysis |
| NPV | negative predictive values |
| RoI | region of interest |
| SVM | support vector machine |
| DICE | similarity index of two images/samples |
| SIL | squamous epithelial lesions |
| VLIR-UOS | Vlaamse Interuniversitaire Raad Universitaire Ontwikkelingssamenwerking (Flemish Interuniversities Council University Development Co-operation) |
| WHO | World Health Organization |
References
- Bruni, L.B.R.L.; Barrionuevo-Rosas, L.; Albero, G.; Serrano, B.; Mena, M.; Collado, J.J.; Gomez, D.; Munoz, J.; Bosch, F.X.; de Sanjose, S. Human Papillomavirus and Related Diseases Report; ICO/IARC Information Centre on HPV and Cancer (HPV Information Centre): Barcelona, Spain, 2019. [Google Scholar]
- Liu, L.; Wang, Y.; Liu, X.; Han, S.; Jia, L.; Meng, L.; Yang, Z.; Chen, W.; Zhang, Y.; Qiao, X. Computer-aided diagnostic system based on deep learning for classifying colposcopy images. Ann. Transl. Med. 2021, 9, 1045. [Google Scholar] [CrossRef] [PubMed]
- Cordero, F.C.; Ayala, P.C.; Maldonado, J.Y.; Montenegro, W.T. Trends in cancer incidence and mortality over three decades in Quito—Ecuador. Colomb. Med. Cali Colomb. 2018, 49, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today. International Agency for Research on Cancer: Lyon, France. 2020. Available online: https://gco.iarc.fr/today (accessed on 11 February 2022).
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Waggoner, S.E. Cervical cancer. Lancet 2003, 361, 2217–2225. [Google Scholar] [CrossRef]
- Yuan, C.; Yao, Y.; Cheng, B.; Cheng, Y.; Li, Y.; Li, Y.; Liu, X.; Cheng, X.; Xie, X.; Wu, J.; et al. The application of deep learning based diagnostic system to cervical squamous intraepithelial lesions recognition in colposcopy images. Sci. Rep. 2020, 10, 11639. [Google Scholar] [CrossRef]
- Chandran, V.; Sumithra, M.G.; Karthick, A.; George, T.; Deivakani, M.; Elakkiya, B.; Subramaniam, U.; Manoharan, S. Diagnosis of Cervical Cancer based on Ensemble Deep Learning Network using Colposcopy Images. BioMed Res. Int. 2021, 2021, 5584004. [Google Scholar] [CrossRef]
- Cho, B.-J.; Choi, Y.J.; Lee, M.-J.; Kim, J.H.; Son, G.-H.; Park, S.-H.; Kim, H.-B.; Joo, Y.-J.; Cho, H.-Y.; Kyung, M.S.; et al. Classification of cervical neoplasms on colposcopic photography using deep learning. Sci. Rep. 2020, 10, 13652. [Google Scholar] [CrossRef]
- Zhang, T.; Luo, Y.-M.; Li, P.; Liu, P.-Z.; Du, Y.-Z.; Sun, P.; Dong, B.; Xue, H. Cervical precancerous lesions classification using pre-trained densely connected convolutional networks with colposcopy images. Biomed. Signal Process. Control 2020, 55, 101566. [Google Scholar] [CrossRef]
- Miyagi, Y.; Takehara, K.; Miyake, T. Application of deep learning to the classification of uterine cervical squamous epithelial lesion from colposcopy images. Mol. Clin. Oncol. 2019, 11, 583–589. [Google Scholar] [CrossRef] [Green Version]
- Sato, M.; Horie, K.; Hara, A.; Miyamoto, Y.; Kurihara, K.; Tomio, K.; Yokota, H. Application of deep learning to the classification of images from colposcopy. Oncol. Lett. 2018, 15, 3518–3523. [Google Scholar] [CrossRef] [Green Version]
- Crespo, B.V.; Neira, V.A.; Segarra, J.O.; Rengel, R.M.; López, D.; Orellana, M.P.; Gómez, A.; Vicuña, M.J.; Mejía, J.; Benoy, I.; et al. Role of Self-Sampling for Cervical Cancer Screening: Diagnostic Test Properties of Three Tests for the Diagnosis of HPV in Rural Communities of Cuenca, Ecuador. Int. J. Environ. Res. Public Health 2022, 19, 4619. [Google Scholar] [CrossRef] [PubMed]
- Motamed, S.; Rogalla, P.; Khalvati, F. Data Augmentation using Generative Adversarial Networks (GANs) for GAN-based Detection of Pneumonia and COVID-19 in Chest X-ray Images. arXiv 2021, arXiv:2006.03622. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Wu, X.; Luo, C.; Ren, P. Deep learning in remote sensing scene classification: A data aug-mentation enhanced convolutional neural network framework. GISci. Remote Sens. 2017, 54, 741–758. [Google Scholar] [CrossRef] [Green Version]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional Networks for Biomedical Image Segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Herz, C.; Fillion-Robin, J.-C.; Onken, M.; Riesmeier, J.; Lasso, A.; Pinter, C.; Fichtinger, G.; Pieper, S.; Clunie, D.; Kikinis, R.; et al. DCMQI: An Open Source Library for Standardized Communication of Quantitative Image Analysis Results Using DICOM. Cancer Res. 2017, 77, e87–e90. [Google Scholar] [CrossRef] [Green Version]
- Fedorov, A.; Clunie, D.; Ulrich, E.; Bauer, C.; Wahle, A.; Brown, B.; Onken, M.; Riesmeier, J.; Pieper, S.; Kikinis, R.; et al. DICOM for quantitative imaging biomarker development: A standards based approach to sharing clinical data and structured PET/CT analysis results in head and neck cancer research. PeerJ 2016, 4, e2057. [Google Scholar] [CrossRef]
- Mayerhoefer, M.E.; Materka, A.; Langs, G.; Häggström, I.; Szczypiński, P.; Gibbs, P.; Cook, G. Introduction to radiomics. J. Nucl. Med. 2020, 61, 488–495. [Google Scholar] [CrossRef]
- Boser, B.E.; Guyon, I.M.; Vapnik, V.N. A training algorithm for optimal margin classifiers. In Proceedings of the Fifth Annual Workshop on Computational Learning Theory, Pittsburgh, PA, USA, 27–29 July 1992; pp. 144–152. [Google Scholar]
- Castillo, D.; Lakshminarayanan, V.; Rodríguez-Álvarez, M.J. MR images, brain lesions, and deep learning. Appl. Sci. 2021, 11, 1675. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, S.-G. Cervical image classification based on image segmentation preprocessing and a CapsNet network model. Int. J. Imaging Syst. Technol. 2019, 29, 19–28. [Google Scholar] [CrossRef] [Green Version]
- Elayaraja, P.; Suganthi, M. Automatic approach for cervical cancer detection and segmentation using neural network classifier. Asian Pac. J. Cancer Prev. APJCP 2018, 19, 3571. [Google Scholar]
- Mehlhorn, G.; Münzenmayer, C.; Benz, M.; Kage, A.; Beckmann, M.W.; Wittenberg, T. Computer-assisted diagnosis in colposcopy: Results of a preliminary experiment? Acta Cytol. 2012, 56, 554–559. [Google Scholar] [CrossRef] [Green Version]
- Acosta-Mesa, H.G.; Cruz-Ramírez, N.; Hernández-Jiménez, R. Acetowhite temporal pattern classification using k-NN to identify precancerous cervical lesion in colposcopic images. Comput. Biol. Med. 2009, 39, 778–784. [Google Scholar] [CrossRef] [PubMed]
- Ragab, D.A.; Sharkas, M.; Marshall, S.; Ren, J. Breast cancer detection using deep convolutional neural networks and support vector machines. PeerJ 2019, 7, e6201. [Google Scholar] [CrossRef] [PubMed]
- Karimi-Zarchi, M.; Zanbagh, L.; Shafii, A.; Taghipour-Zahir, S.; Teimoori, S.; Yazdian-Anari, P. Comparison of pap smear and colposcopy in screening for cervical cancer in patients with secondary immuno-deficiency. Electron. Phys. 2015, 7, 1542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barut, M.U.; Kale, A.; Kuyumcuoğlu, U.; Bozkurt, M.; Ağaçayak, E.; Özekinci, S.; Gül, T. Analysis of Sensitivity, Specificity, and Positive and Negative Predictive Values of Smear and Colposcopy in Diagnosis of Premalignant and Malignant Cervical Lesions. Med Sci. Monit. Int. Med. J. Exp. Clin. Res. 2015, 21, 3860. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. Unet++: A Nested u-net Architecture for Medical Image Segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Springer: Cham, Switzerland, 2018; pp. 3–11. [Google Scholar]
- Liu, J.; Chen, Q.; Fan, J.; Wu, Y. HSIL Colposcopy Image Segmentation Using Improved U-Net. In Proceedings of the 2021 36th Youth Academic Annual Conference of Chinese Association of Automation (YAC), Nanchang, China, 28–30 May 2021; pp. 891–897. [Google Scholar]
- Liu, Y.; Bai, B.; Chen, H.C.; Liu, P.; Feng, H.M. Cervical image segmentation using U-Net model. In Proceedings of the 2019 International Symposium on Intelligent Signal Processing and Communication Systems (ISPACS), Taipei, Taiwan, 3–6 December 2019; pp. 1–2. [Google Scholar]
- Soulami, K.B.; Kaabouch, N.; Saidi, M.N.; Tamtaoui, A. Breast cancer: One-stage automated detection, segmentation, and classification of digital mammograms using UNet model based-semantic segmentation. Biomed. Signal Process. Control 2021, 66, 102481. [Google Scholar] [CrossRef]
- Qi, C.; Chen, J.; Xu, G.; Xu, Z.; Lukasiewicz, T.; Liu, Y. SAG-GAN: Semi-Supervised Attention-Guided GANs for Data Augmentation on Medical Images. arXiv 2020, arXiv:2011.07534. [Google Scholar]
- Negi, A.; Raj, A.N.J.; Nersisson, R.; Zhuang, Z.; Murugappan, M. RDA-UNET-WGAN: An Accurate Breast Ultrasound Lesion Segmentation Using Wasserstein Generative Adversarial Networks. Arab. J. Sci. Eng. 2020, 45, 6399–6410. [Google Scholar] [CrossRef]
- Liang, M.; Zheng, G.; Huang, X.; Milledge, G.; Tokuta, A. Identification of abnormal cervical regions from colposcopy image sequences; UNION Agency: 2013. Available online: https://dspace5.zcu.cz/handle/11025/10655 (accessed on 2 June 2022).
- Jiménez-Gaona, Y.; Rodríguez-Álvarez, M.J.; Lakshminarayanan, V. Deep-Learning-Based Computer-Aided Systems for Breast Cancer Imaging: A Critical Review. Appl. Sci. 2020, 10, 8298. [Google Scholar] [CrossRef]
- Thohir, M.; Foeady, A.Z.; Novitasari, D.C.R.; Arifin, A.Z.; Phiadelvira, B.Y.; Asyhar, A.H. Classification of colposcopy data using GLCM-SVM on cervical cancer. In Proceedings of the 2020 International Conference on Artificial Intelligence in Information and Communication (ICAIIC), Fukuoka, Japan, 19–21 February 2020; pp. 373–378. [Google Scholar]
- Park, Y.R.; Kim, Y.J.; Ju, W.; Nam, K.; Kim, S.; Kim, K.G. Comparison of machine and deep learning for the classification of cervical cancer based on cervicography images. Sci. Rep. 2021, 11, 16143. [Google Scholar] [CrossRef]








| Datasets | Real Images | ||
|---|---|---|---|
| Negative | Positive | Total | |
| Intel & Mobile ODT Cervical Cancer Screening (public) | 130 | 130 | 360 |
| CAMIE (private) | 6 | 14 | 20 |
| Data augmentation | 50 | 50 | 100 |
| Total | 236 | 244 | 480 |
| Hyperparameters | Unet |
|---|---|
| Number of epochs | 200 |
| Batch size | 3 |
| Steps | 123 |
| Steps validation | 30 |
| Optimizer | Adam |
| Learning rate | 0.0005 |
| Loss validation | 0.63 |
| Loss function | Binary-cross entropy |
| Activation function | ReLu, Sigmoid |
| Mean Paired Samples | 95% Confidence Interval | p Value | |
|---|---|---|---|
| Colpo-Experts | 70 | 48–92 | 0.597 |
| Neural Network | 71 | 54–96 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiménez Gaona, Y.; Castillo Malla, D.; Vega Crespo, B.; Vicuña, M.J.; Neira, V.A.; Dávila, S.; Verhoeven, V. Radiomics Diagnostic Tool Based on Deep Learning for Colposcopy Image Classification. Diagnostics 2022, 12, 1694. https://doi.org/10.3390/diagnostics12071694
Jiménez Gaona Y, Castillo Malla D, Vega Crespo B, Vicuña MJ, Neira VA, Dávila S, Verhoeven V. Radiomics Diagnostic Tool Based on Deep Learning for Colposcopy Image Classification. Diagnostics. 2022; 12(7):1694. https://doi.org/10.3390/diagnostics12071694
Chicago/Turabian StyleJiménez Gaona, Yuliana, Darwin Castillo Malla, Bernardo Vega Crespo, María José Vicuña, Vivian Alejandra Neira, Santiago Dávila, and Veronique Verhoeven. 2022. "Radiomics Diagnostic Tool Based on Deep Learning for Colposcopy Image Classification" Diagnostics 12, no. 7: 1694. https://doi.org/10.3390/diagnostics12071694
APA StyleJiménez Gaona, Y., Castillo Malla, D., Vega Crespo, B., Vicuña, M. J., Neira, V. A., Dávila, S., & Verhoeven, V. (2022). Radiomics Diagnostic Tool Based on Deep Learning for Colposcopy Image Classification. Diagnostics, 12(7), 1694. https://doi.org/10.3390/diagnostics12071694