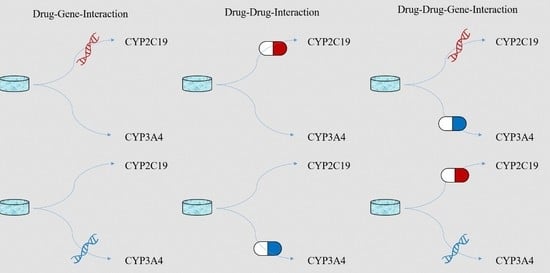
Impact of Drug-Gene-Interaction, Drug-Drug-Interaction, and Drug-Drug-Gene-Interaction on (es)Citalopram Therapy: The PharmLines Initiative
Abstract
:1. Introduction
2. Methods
2.1. Study Design, Setting and Data Sources
2.2. Study Population
2.3. Genotyping
2.4. Definition of Exposures
2.5. Study Outcomes
2.6. Co-Variates
2.7. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Abbing-Karahagopian, V.; Huerta, C.; Souverein, P.C.; De Abajo, F.; Leufkens, H.G.M.; Slattery, J.; Alvarez, Y.; Miret, M.; Gil, M.; Oliva, B.; et al. Antidepressant prescribing in five European countries: Application of common definitions to assess the prevalence, clinical observations, and methodological implications. Eur. J. Clin. Pharmacol. 2014, 70, 849–857. [Google Scholar] [CrossRef]
- Kaplan, C.; Zhang, Y. Assessing the comparative-effectiveness of antidepressants commonly prescribed for depression in the US Medicare population. J. Ment. Health Policy Econ. 2012, 15, 171–178. [Google Scholar]
- Li, G.; Shen, Y.; Luo, J.; Li, H. Efficacy of escitalopram monotherapy in the treatment of major depressive disorder: A pooled analysis of 4 Chinese clinical trials. Medicine 2017, 96, e8142. [Google Scholar] [CrossRef]
- Trivedi, M.H.; Rush, A.J.; Wisniewski, S.R.; Nierenberg, A.A.; Warden, D.; Ritz, L.; Norquist, G.; Howland, R.H.; Lebowitz, B.; McGrath, P.J.; et al. Evaluation of outcomes with citalopram for depression using measurement-based care in STAR* D: Implications for clinical practice. Am. J. Psychiatry 2006, 163, 28–40. [Google Scholar] [CrossRef]
- Fredricson Overo, K. Kinetics of citalopram in man; plasma levels in patients. Prog. Neuropsychopharmacol. Biol. Psychiatry 1982, 6, 311–318. [Google Scholar] [CrossRef]
- Jin, Y.; Pollock, B.G.; Frank, E.; Cassano, G.B.; Rucci, P.; Müller, D.J.; Kennedy, J.L.; Forgione, R.N.; Kirshner, M.; Kepple, G.; et al. Effect of age, weight, and CYP2C19 genotype on escitalopram exposure. J. Clin. Pharmacol. 2010, 50, 62–72. [Google Scholar] [CrossRef] [Green Version]
- Chang, M.; Tybring, G.; Dahl, M.L.; Lindh, J.D. Impact of cytochrome P450 2C19 polymorphisms on citalopram/escitalopram exposure: A systematic review and meta-analysis. Clin. Pharmacokinet. 2014, 53, 801–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rochat, B.; Amey, M.; Gillet, M.; Meyer, U.A.; Baumann, P. Identification of three cytochrome P450 isozymes involved in N-demethylation of citalopram enantiomers in human liver microsomes. Pharmacogenetics 1997, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- von Moltke, L.L.; Greenblatt, D.J.; Giancarlo, G.M.; Granda, B.W.; Harmatz, J.S.; Shader, R.I. Escitalopram (S-citalopram) and its metabolites in vitro: Cytochromes mediating biotransformation, inhibitory effects, and comparison to R-citalopram. Drug Metab. Dispos. 2001, 29, 1102–1109. [Google Scholar] [PubMed]
- Fudio, S.; Borobia, A.M.; Piñana, E.; Ramírez, E.; Tabarés, B.; Guerra, P.; Carcas, A.; Frías, J. Evaluation of the influence of sex and CYP2C19 and CYP2D6 polymorphisms in the disposition of citalopram. Eur. J. Pharmacol. 2010, 626, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Westervelt, P.; Cho, K.; Bright, D.R.; Kisor, D.F. Drug-gene interactions: Inherent variability in drug maintenance dose requirements. Pharm. Ther. 2014, 39, 630–637. [Google Scholar]
- Wenzel-Seifert, K.; Brandl, R.; Hiemke, C.; Haen, E. Influence of concomitant medications on the total clearance and the risk for supra-therapeutic plasma concentrations of Citalopram. A population-based cohort study. Pharmacopsychiatry 2014, 47, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Thirumaran, R.K.; Heck, J.W.; Hocum, B.T. CYP450 genotyping and cumulative drug–gene interactions: An update for precision medicine. Future Med. 2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verbeurgt, P.; Mamiya, T.; Oesterheld, J. How common are drug and gene interactions? Prevalence in a sample of 1143 patients with CYP2C9, CYP2C19 and CYP2D6 genotyping. Pharmacogenomics 2014, 15, 655–665. [Google Scholar] [CrossRef] [PubMed]
- Bahar, M.A.; Setiawan, D.; Hak, E.; Wilffert, B. Pharmacogenetics of drug–drug interaction and drug–drug–gene interaction: A systematic review on CYP2C9, CYP2C19 and CYP2D6. Pharmacogenomics 2017, 18, 701–739. [Google Scholar] [CrossRef] [PubMed]
- Malki, M.A.; Pearson, E.R. Drug–drug–gene interactions and adverse drug reactions. Pharm. J. 2019, 20, 355–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hicks, J.K.; Bishop, J.R.; Sangkuhl, K.; Müller, D.J.; Ji, Y.; Leckband, S.G.; Leeder, J.S.; Graham, R.L.; Chiulli, D.L.; LLerena, A.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin. Pharmacol. Ther. 2015, 98, 127–134. [Google Scholar] [CrossRef] [Green Version]
- Swen, J.J.; Nijenhuis, M.; de Boer, A.; Grandia, L.; Maitland-van der Zee, A.H.; Mulder, H.; Rongen, G.A.P.J.M.; Van Schaik, R.H.N.; Schalekamp, T.; Touw, D.J.; et al. Pharmacogenetics: From bench to byte—An update of guidelines. Clin. Pharmacol. Ther. 2011, 89, 662–673. [Google Scholar] [CrossRef]
- Sediq, R.; van der Schans, J.; Dotinga, A.; Alingh, R.A.; Wilffert, B.; Bos, J.H.; Schuiling-Veninga, C.C.; Hak, E. Concordance assessment of self-reported medication use in the netherlands three-generation lifelines Cohort study with the pharmacy database iaDB. nl: The Pharmlines initiative. Clin. Epidemiol. 2018, 10, 981. [Google Scholar] [CrossRef] [Green Version]
- Jukić, M.M.; Haslemo, T.; Molden, E.; Ingelman-Sundberg, M. Impact of CYP2C19 genotype on escitalopram exposure and therapeutic failure: A retrospective study based on 2087 patients. Am. J. Psychiatry 2018, 175, 463–470. [Google Scholar] [CrossRef] [Green Version]
- Bijl, M.J.; Visser, L.E.; Hofman, A.; Vulto, A.G.; Van Gelder, T.; Stricker, B.H.C.; Van Schaik, R.H. Influence of the CYP2D6* 4 polymorphism on dose, switching and discontinuation of antidepressants. Br. J. Clin. Pharmacol. 2008, 65, 558–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scholtens, S.; Smidt, N.; Swertz, M.A.; Bakker, S.J.; Dotinga, A.; Vonk, J.M.; Van Dijk, F.; van Zon, S.K.; Wijmenga, C.; Wolffenbuttel, B.H.; et al. Cohort Profile: LifeLines, a three-generation cohort study and biobank. Int. J. Epidemiol. 2014, 44, 1172–1180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stolk, R.P.; Rosmalen, J.G.; Postma, D.S.; de Boer, R.A.; Navis, G.; Slaets, J.P.; Ormel, J.; Wolffenbuttel, B.H. Universal risk factors for multifactorial diseases. Eur. J. Epidemiol. 2008, 23, 67–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klijs, B.; Scholtens, S.; Mandemakers, J.J.; Snieder, H.; Stolk, R.P.; Smidt, N. Representativeness of the LifeLines cohort study. PLoS ONE 2015, 10, e0137203. [Google Scholar] [CrossRef]
- Visser, S.T.; Schuiling-Veninga, C.C.; Bos, J.H.; de Jong-van den Berg Lolkje, T.W.; Postma, M.J. The population-based prescription database IADB. nl: Its development, usefulness in outcomes research and challenges. Expert Rev. Pharm. Outcomes Res. 2013, 13, 285–292. [Google Scholar]
- Bahar, M.; Hak, E.; Bos, J.H.; Borgsteede, S.D.; Wilffert, B. The burden and management of cytochrome P450 2D6 (CYP2D6)-mediated drug–drug interaction (DDI): Co-medication of metoprolol and paroxetine or fluoxetine in the elderly. Pharmacoepidemiol. Drug Saf. 2017, 26, 752–765. [Google Scholar] [CrossRef]
- Bahar, M.A.; Wang, Y.; Bos, J.H.; Wilffert, B.; Hak, E. Discontinuation and dose adjustment of metoprolol after metoprolol-paroxetine/fluoxetine co-prescription in Dutch elderly. Pharmacoepidemiol. Drug Saf. 2018, 27, 621–629. [Google Scholar] [CrossRef] [Green Version]
- Daud, A.N.; Bergman, J.E.; Oktora, M.P.; Kerstjens-Frederikse, W.S.; Groen, H.; Bos, J.H.; Hak, E.; Wilffert, B. Maternal use of drug substrates of placental transporters and the effect of transporter-mediated drug interactions on the risk of congenital anomalies. PLoS ONE 2017, 12, e0173530. [Google Scholar] [CrossRef]
- Gaedigk, A.; Ingelman-Sundberg, M.; Miller, N.A.; Leeder, J.S.; Whirl-Carrillo, M.; Klein, T.E.; PharmVar Steering Committee. The Pharmacogene Variation (PharmVar) Consortium: Incorporation of the human cytochrome P450 (CYP) allele nomenclature database. Clin. Pharmacol. Ther. 2018, 103, 399–401. [Google Scholar] [CrossRef] [Green Version]
- García-Martín, E.; Martínez, C.; Pizarro, R.M.; García-Gamito, F.J.; Gullsten, H.; Raunio, H.; Agúndez, J.A. CYP3A4 variant alleles in white individuals with low CYP3A4 enzyme activity. Clin. Pharmacol. Ther. 2002, 71, 196–204. [Google Scholar] [CrossRef]
- Zhang, W.; Chang, Y.Z.; Kan, Q.C.; Zhang, L.R.; Li, Z.S.; Lu, H.; Wang, Z.Y.; Chu, Q.J.; Zhang, J. CYP3A4* 1G genetic polymorphism influences CYP3A activity and response to fentanyl in Chinese gynecologic patients. Eur. J. Clin. Pharmacol. 2010, 66, 61. [Google Scholar] [CrossRef] [PubMed]
- Borgsteede, S. Commentaren Medicatiebewaking; Health Base: Houten, The Netherlands, 2015. [Google Scholar]
- Flockhart, D. Drug Interactions: Cytochrome P450 Drug Interaction Table; Indiana University School of Medicine: Indianapolis, IN, USA, 2012. [Google Scholar]
- Palaniyappan, L.; Insole, L.; Ferrier, N. Combining antidepressants: A review of evidence. Adv. Psychiatr. Treat. 2009, 15, 90–99. [Google Scholar] [CrossRef] [Green Version]
- Olfson, M.; Marcus, S.C.; Tedeschi, M.; Wan, G.J. Continuity of antidepressant treatment for adults with depression in the United States. Am. J. Psychiatry 2006, 163, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Saragoussi, D.; Chollet, J.; Bineau, S.; Chalem, Y.; Milea, D. Antidepressant switching patterns in the treatment of major depressive disorder: A General Practice Research Database (GPRD) study. Int. J. Clin. Pract. 2012, 66, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Hansen, R.A.; Dusetzina, S.B.; Dominik, R.C.; Gaynes, B.N. Prescription refill records as a screening tool to identify antidepressant non-adherence. Pharmacoepidemiol. Drug Saf. 2010, 19, 33–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Boven, J.F.; Van Raaij, J.J.; Van Der Galiën, R.; Postma, M.J.; Van Der Molen, T.; Dekhuijzen, P.R.; Vegter, S. Impact of multiple-dose versus single-dose inhaler devices on COPD patients’ persistence with long-acting β 2-agonists: A dispensing database analysis. NPJ Prim. Care Respir. Med. 2014, 24, 14069. [Google Scholar] [CrossRef] [Green Version]
- De Jong, J.C.; Van Den Berg, P.B.; Tobi, H.; De Jong, L.T.; Van Den Berg. Combined use of SSRIs and NSAIDs increases the risk of gastrointestinal adverse effects. Br. J. Clin. Pharmacol. 2003, 55, 591–595. [Google Scholar] [CrossRef] [Green Version]
- Mrazek, D.A.; Biernacka, J.M.; O’Kane, D.J.; Black, J.L.; Cunningham, J.M.; Drews, M.S.; Snyder, K.A.; Stevens, S.R.; Rush, A.J.; Weinshilboum, R.M. CYP2C19 variation and citalopram response. Pharmacogenet. Genom. 2011, 21, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Dutch Pharmacogenetics Working Group. Dutch Pharmacogenetics Working Group Guidelines; Dutch Pharmacogenetics Working Group: The Hague, The Netherlands, 2018. [Google Scholar]
- Bahar, M.; Bos, J.H.; Borgsteede, S.D.; Dotinga, A.; Alingh, R.A.; Wilffert, B.; Hak, E. Prevalence and Accuracy of Information on CYP2D6, CYP2C19, and CYP2C9 Related Substrate and Inhibitor Co-Prescriptions in the General Population: A Cross-Sectional Descriptive Study as Part of the PharmLines Initiative. Front. Pharmacol. 2020, 11, 624. [Google Scholar] [CrossRef]
- Malling, D.; Poulsen, M.; Søgaard, B. The effect of cimetidine or omeprazole on the pharmacokinetics of escitalopram in healthy subjects. Br. J. Clin. Pharmacol. 2005, 60, 287–290. [Google Scholar] [CrossRef] [Green Version]
- Rocha, A.; Coelho, E.B.; Sampaio, S.A.; Lanchote, V.L. Omeprazole preferentially inhibits the metabolism of (+)-(S)-citalopram in healthy volunteers. Br. J. Clin. Pharmacol. 2010, 70, 43–51. [Google Scholar] [CrossRef] [Green Version]
- Gjestad, C.; Westin, A.A.; Skogvoll, E.; Spigset, O. Effect of proton pump inhibitors on the serum concentrations of the selective serotonin reuptake inhibitors citalopram, escitalopram, and sertraline. Ther. Drug Monit. 2015, 37, 90–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Storelli, F.; Matthey, A.; Lenglet, S.; Thomas, A.; Desmeules, J.; Daali, Y. Impact of CYP2D6 functional allelic variations on phenoconversion and drug–drug interactions. Clin. Pharmacol. Ther. 2018, 104, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Wijnen, P.A.H.M.; Op den Buijsch, R.A.M.; Drent, M.; Kuipers, P.M.J.C.; Neef, C.; Bast, A.; Bekers, O.; Koek, G.H. The prevalence and clinical relevance of cytochrome P450 polymorphisms. Aliment. Pharmacol. Ther. 2007, 26, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Herrlin, K.; Yasui-Furukori, N.; Tybring, G.; Widén, J.; Gustafsson, L.L.; Bertilsson, L. Metabolism of citalopram enantiomers in CYP2C19/CYP2D6 phenotyped panels of healthy Swedes. Br. J. Clin. Pharmacol. 2003, 56, 415–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Haplotype Name | Gene | rsID | Reference Sequence | Variant. Start | Variant. Stop | Reference. Allele | Variant. Allele | Type |
|---|---|---|---|---|---|---|---|---|
| CYP2C19*1 | CYP2C19 | rs3758581 | 10 | 96602622 | 96602622 | G | - | single |
| CYP2C19*1 | CYP2C19 | rs12769205 | 10 | 96535123 | 96535123 | A | - | single |
| CYP2C19*1 | CYP2C19 | rs28399504 | 10 | 96522462 | 96522462 | A | - | single |
| CYP2C19*1 | CYP2C19 | rs41291556 | 10 | 96535172 | 96535172 | T | - | single |
| CYP2C19*1 | CYP2C19 | rs11188072 | 10 | 96519060 | 96519060 | C | - | single |
| CYP2C19*2 | CYP2C19 | rs12769205 | 10 | 96535123 | 96535123 | A | G | single |
| CYP2C19*4 | CYP2C19 | rs28399504 | 10 | 96522462 | 96522462 | A | G | single |
| CYP2C19*5/7 | CYP2C19 | rs3758581 | 10 | 96602622 | 96602622 | G | A | single |
| CYP2C19*8 | CYP2C19 | rs41291556 | 10 | 96535172 | 96535172 | T | C | single |
| CYP2C19*17 | CYP2C19 | rs11188072 | 10 | 96519060 | 96519060 | C | T | single |
| Haplotype. Name | Gene | rsID | Reference Sequence | Variant. Start | Variant. Stop | Reference. Allele | Variant. Allele | Type |
|---|---|---|---|---|---|---|---|---|
| CYP3A4*1A | CYP3A4 | rs2740574 | 7 | 99382095 | 99382095 | T | - | single |
| CYP3A4*1A | CYP3A4 | rs2242480 | 7 | 99361465 | 99361465 | C | - | single |
| CYP3A4*1A | CYP3A4 | rs35599367 | 7 | 99366315 | 99366315 | G | - | single |
| CYP3A4*1B | CYP3A4 | rs2740574 | 7 | 99382095 | 99382095 | T | C | single |
| CYP3A4*1G | CYP3A4 | rs2242480 | 7 | 99361465 | 99361465 | C | T | single |
| CYP3A4*22 | CYP3A4 | rs35599367 | 7 | 99366315 | 99366315 | G | A | single |
| Gene | Haplotype | Metabolic Function | Reference |
|---|---|---|---|
| CYP2C19 | CYP2C19*1 | Normal | [29] |
| CYP2C19*2 | No | [29] | |
| CYP2C19*4 | No | [29] | |
| CYP2C19*5/7 | No | [29] | |
| CYP2C19*8 | No | [29] | |
| CYP2C19*17 | Increased | [29] | |
| CYP3A4 | CYP3A4*1A | Normal | [29] |
| CYP3A4*1B | Normal | [30] | |
| CYP3A4*1G | Decreased | [31] | |
| CYP3A4*22 | Decreased | [29] |
| CYP2C19 | No | Normal | Increased | CYP3A4 | Decreased | Normal |
|---|---|---|---|---|---|---|
| No | PM | IM | IM | Decreased | PM | IM |
| Normal | IM | NM | NM | Normal | IM | NM |
| Increased | IM | NM | UM |
| Variabels | N | % |
|---|---|---|
| Gender (n women, %) | 200 | 63.3 |
| Age in years, median (IQR) | 45 | 14 |
| CYP2C19 Phenotypes | ||
| CYP2C19 NM (n, %) | 176 | 55.7 |
| CYP2C19 IM (n, %) | 103 | 32.6 |
| CYP2C19 PM (n, %) | 23 | 7.3 |
| CYP2C19 UM (n, %) | 14 | 4.4 |
| CYP3A4 Phenotypes | ||
| CYP3A4 NM (n, %) | 254 | 80.4 |
| CYP3A4 IM (n, %) | 56 | 17.7 |
| CYP3A4 PM (n, %) | 6 | 1.9 |
| Combination of CYP2C19 & CYP3A4 Phenotypes | ||
| CYP2C19 NM + CYP3A4 NM (n, %) | 140 | 44.3 |
| CYP2C19 IM/PM + CYP3A4 NM (n, %) | 104 | 32.9 |
| CYP2C19 IM/PM + CYP3A4 IM/PM (n, %) | 20 | 6.3 |
| CYP2C19 NM + CYP3A4 IM/PM (n, %) | 36 | 11.4 |
| CYP2C19 UM + CYP3A4 NM/IM (n, %) | 14 | 4.4 |
| Type of CYP modulator combination | ||
| No inhibitor or inducer of CYP2C19/3A4/2D6 | 260 | 82.3 |
| CYP2C19 inhibitor alone (n, %) | 44 | 13.9 |
| CYP3A4 inhibitor alone (n, %) | 4 | 1.3 |
| CYP2D6 inhibitor alone (n, %) | 6 | 1.9 |
| CYP2C19 inhibitor + CYP2D6 inhibitor (n, %) * | 1 | 0.3 |
| CYP2C19 inhibitor + CYP3A4 inducer (n, %) * | 1 | 0.3 |
| DDD at start of citalopram and escitalopram | ||
| DDD < 1 (n, %) | 25 | 7.9 |
| DDD >= 1 (n, %) | 197 | 62.3 |
| No dose information (n, %) | 94 | 29.7 |
| Potential comorbidities | ||
| No comorbidity (n, %) | 65 | 20.6 |
| 1–2 potential comorbidities (n, %) | 216 | 68.3 |
| ≥3 potential comorbidities (n, %) | 35 | 11.1 |
| Number of co-prescriptions during (es)citalopram | ||
| 1–3 type of drugs (n, %) | 247 | 78.2 |
| >3 type of drugs (n, %) | 69 | 21.8 |
| Number of CYP modulator during (es)citalopram | ||
| No CYP modulator (n, %) | 260 | 82.3 |
| 1 CYP modulator (n, %) | 27 | 8.5 |
| ≥2 CYP modulator (n, %) | 29 | 9.2 |
| Combined exposures | ||
| No exposures | ||
| CYP2C19 NM + CYP3A4 NM + No CYP Modulator (n, %) | 111 | 35.1 |
| DDI | ||
| CYP2C19 NM + CYP3A4 NM + Yes CYP Modulator (n, %) | 29 | 9.2 |
| DGI | ||
| CYP2C19 IM/PM + CYP3A4 NM + No CYP Modulator (n, %) | 89 | 28.2 |
| CYP2C19 IM/PM + CYP3A4 IM/PM + No CYP Modulator (n, %) | 20 | 6.3 |
| CYP2C19 NM + CYP3A4 IM/PM + No CYP Modulator (n, %) | 29 | 9.2 |
| CYP2C19 UM + CYP3A4 NM/IM + No CYP Modulator (n, %) | 11 | 3.5 |
| DDGI (n, %) | 27 | 8.5 |
| CYP2C19 Phenotype | CYP3A4 Phenotype | CYP2C19 Inhibitor | CYP3A4 Inhibitor | CYP2D6 Inhibitor | CYP2C19 Inducer | CYP3A4 Inducer | N | % |
|---|---|---|---|---|---|---|---|---|
| One pathway | ||||||||
| UM/IM/PM | NM | Y | N | N | N | N | 14 | 51.8 |
| Two pathways | ||||||||
| IM | IM | Y | N | N | N | N | 2 | 7.4 |
| IM | NM | N | Y | N | N | N | 2 | 7.4 |
| IM | NM | N | N | Y | N | N | 2 | 7.4 |
| NM | IM/PM | Y | N | N | N | N | 6 | 22.2 |
| NM | IM | N | N | Y | N | N | 1 | 3.7 |
| SUM | 27 | |||||||
| Variables | Switching * | p-Value | Decreased Dose # | p-Value | Increased Dose # | p-Value | Discontinuation * | p-Value | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yes (n = 25) | No (n = 279) | Yes (n = 7) | No (n = 213) | Yes (n = 80) | No (n = 140) | Yes (n = 47) | No (n = 257) | |||||
| Gender (n Women) | 15 | 177 | 0.73 | 5 | 133 | 1.00 | 40 | 98 | 0.003 | 29 | 163 | 0.82 |
| Age in years (median, IQR) | 41 | 45 | 0.68 | 39 | 42 | 0.38 | 43.5 | 41 | 0.92 | 48 | 44 | 0.03 |
| DDD at start (n DDD ≥1) | 18 | 177 | 1.00 | 7 | 188 | 1.00 | 64 | 131 | 0.002 | 31 | 164 | 0.57 |
| Potential comorbidities (n Yes) | ||||||||||||
| No comorbidity (n) | 3 | 60 | 0.18 | 3 | 36 | 0.22 | 16 | 23 | 0.79 | 9 | 54 | 0.71 |
| 1–2 potential comorbidities (n) | 21 | 185 | 4 | 152 | 55 | 101 | 34 | 172 | ||||
| ≥3 potential comorbidities (n) | 1 | 34 | 0 | 25 | 9 | 16 | 4 | 31 | ||||
| N of co-prescriptions | ||||||||||||
| 1–3 (n) | 24 | 213 | 0.02 | 7 | 166 | 0.35 | 63 | 110 | 0.97 | 38 | 199 | 0.60 |
| >3 (n) | 1 | 66 | 0 | 47 | 17 | 30 | 9 | 58 | ||||
| N of CYP modulator prescriptions | ||||||||||||
| No (n) | 22 | 229 | 0.92 | 6 | 177 | 0.73 | 69 | 114 | 0.62 | 41 | 210 | 0.64 |
| 1 (n) | 2 | 25 | 1 | 18 | 5 | 14 | 4 | 23 | ||||
| ≥2 (n) | 1 | 25 | 0 | 18 | 6 | 12 | 2 | 24 | ||||
| Variables | Switching and/or Dose Reduction | Univariate Analysis | Multivariate Analysis | |||||
|---|---|---|---|---|---|---|---|---|
| Yes (n = 31, %) | No (n = 273, %) | OR (95%CI) | p-Value | q-Value | aOR (95%CI) | p-Value | q-Value | |
| CYP2C19 & CYP3A4 predicted phenotypes | ||||||||
| CYP2C19 predicted phenotypes * | ||||||||
| CYP2C19 NM | 12 (38.7) | 157 (57.5) | Ref. | Ref. | ||||
| CYP2C19 IM | 18 (58.1) | 82 (30) | 2.87 (1.32–6.25) | 0.01 | 0.08 | 3.16 (1.41–7.09) | 0.005 | 0.06 |
| CYP2C19 PM | 1 (3.2) | 20 (7.3) | 0.65 (0.08–5.30) | 0.69 | 0.90 | 0.54 (0.07–4.52) | 0.57 | 0.68 |
| CYP2C19 UM | 0 (0) | 14 (5.1) | NA | NA | ||||
| CYP3A4 predicted phenotypes ** | ||||||||
| CYP3A4 NM | 23 (74.2) | 220 (80.6) | Ref. | |||||
| CYP3A4 IM | 8(25.8) | 47 (17.2) | 1.63 (0.69–3.86) | 0.27 | 0.54 | 1.37 (0.55–3.39) | 0.50 | 0.67 |
| CYP3A4 PM | 0 (0) | 6 (2.2) | NA | NA | ||||
| Combination of predicted phenotypes *** | ||||||||
| CYP2C19 NM + CYP3A4 NM | 9 (29) | 125 (45.8) | Ref. | Ref. | ||||
| CYP2C19 IM/PM + CYP3A4 NM | 14 (45.2) | 85 (31.1) | 2.29 (0.95–5.52) | 0.07 | 0.17 | 2.35 (0.96–5.76) | 0.06 | 0.14 |
| CYP2C19 IM/PM + CYP3A4 IM/PM | 5 (16.1) | 17 (6.2) | 4.08 (1.22–13.63) | 0.02 | 0.08 | 3.46 (1.02–11.75) | 0.05 | 0.14 |
| CYP2C19 NM + CYP3A4 IM/PM | 3 (9.7) | 32 (11.7) | 1.30 (0.33–5.09) | 0.70 | 0.90 | 1.11 (0.28–4.43) | 0.88 | 0.96 |
| CYP2C19 UM + CYP3A4 NM/IM | 0 (0) | 14 (5.1) | NA | NA | ||||
| CYP modulator # | ||||||||
| No inhibitor/inducer of CYP2C19/3A4/2D6 | 27 (87.1) | 224 (82.1) | Ref. | Ref. | ||||
| CYP2C19 inhibitor | 4 (12.9) | 40 (14.7) | 0.83 (0.27–2.49) | 0.74 | 0.90 | 2.36 (0.67–8.32) | 0.18 | 0.36 |
| CYP3A4 inhibitor | 0 (0) | 4 (1.5) | NA | NA | ||||
| CYP2D6 inhibitor | 0 (0) | 5 (1.8) | NA | NA | ||||
| Combined exposures ^ | ||||||||
| No exposures | ||||||||
| CYP2C19 NM + CYP3A4 NM + No CYP Modulator | 7 (22.6) | 101 (37) | Ref. | Ref. | ||||
| DDI | ||||||||
| CYP2C19 NM + CYP3A4 NM + Yes CYP Modulator | 2 (6.5) | 24 (8.8) | 1.20 (0.24–6.16) | 0.83 | 0.90 | 2.82 (0.49–15.97) | 0.24 | 0.41 |
| DGI | ||||||||
| CYP2C19 IM/PM + CYP3A4 NM + No CYP Modulator | 13 (41.9) | 71 (26) | 2.64 (1.00–6.95) | 0.05 | 0.15 | 2.75 (1.03–7.29) | 0.04 | 0.14 |
| CYP2C19 IM/PM + CYP3A4 IM/PM + No CYP Modulator | 5 (16.1) | 15 (5.5) | 4.81 (1.35–17.12) | 0.02 | 0.08 | 4.38 (1.22–15.69) | 0.02 | 0.12 |
| CYP2C19 NM + CYP3A4 IM/PM + No CYP Modulator | 2 (6.5) | 26 (9.5) | 1.11 (0.22–5.66) | 0.90 | 0.90 | 1.02 (0.19–5.24) | 0.98 | 0.98 |
| CYP2C19 UM + CYP3A4 NM/IM + No CYP Modulator | 0 (0) | 11 (4) | NA | NA | ||||
| DDGI | 2 (6.5) | 25 (9.2) | 1.15 (0.23–5.89) | 0.86 | 0.90 | 2.33 (0.42–12.78) | 0.33 | 0.49 |
| Variables | Early Discontinuation | Univariate Analysis | Multivariate Analysis | |||||
|---|---|---|---|---|---|---|---|---|
| Yes (n = 47, %) | No (n = 257, %) | OR (95%CI) | p-Value | q-Value | aOR (95%CI) | p-Value | q-Value | |
| CYP2C19 & CYP3A4 predicted phenotypes | ||||||||
| CYP2C19 phenotypes * | ||||||||
| CYP2C19 NM | 33 (70.2) | 136 (52.9) | Ref. | Ref. | ||||
| CYP2C19 IM | 9 (19.1) | 91 (35.4) | 0.41 (0.19–0.89) | 0.03 | 0.45 | 0.35 (0.15–0.79) | 0.01 | 0.15 |
| CYP2C19 PM | 2 (4.3) | 19 (7.4) | 0.43 (0.09–1.96) | 0.28 | 0.50 | 0.41 (0.09–1.89) | 0.25 | 0.54 |
| CYP2C19 UM | 3 (6.4) | 11 (4.3) | 1.12 (0.29–4.26) | 0.86 | 0.86 | 1.24 (0.32–4.88) | 0.75 | 0.75 |
| CYP3A4 phenotypes ** | ||||||||
| CYP3A4 NM | 36 (76.6) | 207 (80.5) | Ref. | Ref. | ||||
| CYP3A4 IM | 11 (23.4) | 44 (17.1) | 1.44 (0.68–3.04) | 0.34 | 0.51 | 1.29 (0.59–2.84) | 0.51 | 0.59 |
| CYP3A4 PM | 0 (0) | 6 (2.3) | NA | NA | ||||
| Combination of predicted phenotypes *** | ||||||||
| CYP2C19 NM + CYP3A4 NM | 24 (51.1) | 110 (42.8) | Ref. | Ref. | ||||
| CYP2C19 IM/PM + CYP3A4 NM | 10 (21.3) | 89 (34.6) | 0.52 (0.23–1.13) | 0.09 | 0.45 | 0.45 (0.20–1.02) | 0.06 | 0.35 |
| CYP2C19 IM/PM + CYP3A4 IM/PM | 1 (2.1) | 21 (8.2) | 0.22 (0.03–1.70) | 0.15 | 0.45 | 0.17 (0.02–1.39) | 0.10 | 0.36 |
| CYP2C19 NM + CYP3A4 IM/PM | 9 (19.1) | 26 (10.1) | 1.59 (0.66–3.81) | 0.30 | 0.50 | 1.43 (0.58–3.53) | 0.44 | 0.59 |
| CYP2C19 UM + CYP3A4 NM/IM | 3 (6.4) | 11 (4.3) | 1.25 (0.32–4.83) | 0.75 | 0.80 | 1.43 (0.36–5.69) | 0.61 | 0.65 |
| CYP modulator # | ||||||||
| No inhibitor/inducer of CYP2C19/3A4/2D6 | 41 (87.2) | 210 (81.7) | Ref. | Ref. | ||||
| CYP2C19 inhibitor alone | 6 (12.8) | 38 (14.8) | 0.81 (0.32–2.04) | 0.65 | 0.80 | 0.68 (0.26–1.75) | 0.42 | 0.59 |
| CYP3A4 inhibitor alone | 0 (0) | 4 (1.6) | NA | NA | ||||
| CYP2D6 inhibitor alone | 0 (0) | 5 (1.9) | NA | NA | ||||
| Combined exposures ^ | ||||||||
| No exposures | ||||||||
| CYP2C19 NM + CYP3A4 NM + No CYP Modulator | 20 (42.6) | 88 (34.2) | Ref. | Ref. | ||||
| DDI | ||||||||
| CYP2C19 NM + CYP3A4 NM + Yes CYP Modulator | 4 (8.5) | 22 (8.6) | 0.80 (0.25–2.58) | 0.71 | 0.80 | 0.67 (0.20–2.21) | 0.51 | 0.59 |
| DGI | ||||||||
| CYP2C19 IM/PM + CYP3A4 NM + No CYP Modulator | 9 (19.1) | 75 (29.2) | 0.53 (0.23–1.23) | 0.14 | 0.45 | 0.44 (0.19–1.06) | 0.07 | 0.35 |
| CYP2C19 IM/PM + CYP3A4 IM/PM + No CYP Modulator | 1 (2.1) | 19 (7.4) | 0.23 (0.03–1.83) | 0.17 | 0.45 | 0.19 (0.02–1.53) | 0.12 | 0.36 |
| CYP2C19 NM + CYP3A4 IM/PM + No CYP Modulator | 8 (17) | 20 (7.8) | 1.76 (0.68–4.56) | 0.25 | 0.50 | 1.52 (0.57–4.04) | 0.41 | 0.59 |
| CYP2C19 UM + CYP3A4 NM/IM + No CYP Modulator | 3 (6.4) | 8 (3.1) | 1.65 (0.40–6.78) | 0.49 | 0.67 | 1.89 (0.45–8.07) | 0.39 | 0.59 |
| DDGI | 2 (4.3) | 25 (9.7) | 0.35 (0.08–1.61) | 0.18 | 0.45 | 0.38 (0.08–1.75) | 0.21 | 0.53 |
| Variables | Dose Elevation | Univariate Analysis | Multivariate Analysis | |||||
|---|---|---|---|---|---|---|---|---|
| Yes (n = 80, %) | No (n = 140, %) | OR (95%CI) | p-Value | q-Value | aOR (95%CI) | p-Value | q-Value | |
| CYP2C19 & CYP3A4 predicted phenotypes | ||||||||
| CYP2C19 predicted phenotypes * | ||||||||
| CYP2C19 NM | 51 (63.7) | 67 (47.9) | Ref. | Ref. | ||||
| CYP2C19 IM | 23 (28.7) | 56 (40) | 0.54 (0.29–0.99) | 0.05 | 0.45 | 0.59 (0.31–1.12) | 0.11 | 0.54 |
| CYP2C19 PM | 4 (5) | 10 (7.1) | 0.53 (0.16–1.77) | 0.29 | 0.61 | 0.56 (0.16–2.02) | 0.38 | 0.54 |
| CYP2C19 UM | 2 (2.5) | 7 (5) | 0.37 (0.07–1.88) | 0.23 | 0.61 | 0.35 (0.07–1.85) | 0.22 | 0.54 |
| CYP3A4 predicted phenotypes ** | ||||||||
| CYP3A4 NM | 61 (76.3) | 114 (81.4) | Ref. | Ref. | ||||
| CYP3A4 IM | 17 (21.3) | 24 (17.1) | 1.32 (0.66–2.65) | 0.43 | 0.61 | 1.48 (0.70–3.12) | 0.30 | 0.54 |
| CYP3A4 PM | 2 (2.5) | 2 (1.4) | 1.87 (0.26–13.59) | 0.54 | 0.66 | 1.27 (0.15–10.64) | 0.82 | 0.87 |
| Combination of predicted phenotypes *** | ||||||||
| CYP2C19 NM + CYP3A4 NM | 40 (50) | 56 (40) | Ref. | Ref. | ||||
| CYP2C19 IM/PM + CYP3A4 NM | 21 (26.3) | 52 (37.1) | 0.56 (0.29–1.08) | 0.08 | 0.45 | 0.69 (0.35–1.36) | 0.28 | 0.54 |
| CYP2C19 IM/PM + CYP3A4 IM/PM | 6 (7.5) | 14 (10) | 0.60 (0.21–1.69) | 0.34 | 0.61 | 0.57 (0.19–1.68) | 0.31 | 0.54 |
| CYP2C19 NM + CYP3A4 IM/PM | 11 (13.8) | 11 (7.9) | 1.40 (0.55–3.54) | 0.48 | 0.63 | 1.66 (0.62–4.49) | 0.31 | 0.54 |
| CYP2C19 UM + CYP3A4 NM/IM | 2 (2.5) | 7 (5) | 0.40 (0.08–2.03) | 0.27 | 0.61 | 0.41 (0.08–2.18) | 0.29 | 0.54 |
| CYP modulator # | ||||||||
| No inhibitor/inducer of CYP2C19/3A4/2D6 | 69 (86.3) | 114 (81.4) | Ref. | Ref. | ||||
| CYP2C19 inhibitor alone | 9 (11.3) | 21 (15) | 0.71 (0.31–1.63) | 0.42 | 0.61 | 0.80 (0.33–1.95) | 0.63 | 0.76 |
| CYP3A4 inhibitor alone | 2 (2.5) | 2 (1.4) | 1.65 (0.23–11.99) | 0.62 | 0.70 | 2.75 (0.37–20.74) | 0.33 | 0.54 |
| CYP2D6 inhibitor alone | 0 (0) | 3 (2.1) | NA | NA | ||||
| Combined exposures ^ | ||||||||
| No exposures | ||||||||
| CYP2C19 NM + CYP3A4 NM + No CYP Modulator | 33 (41.3) | 46 (32.9) | Ref. | Ref. | ||||
| DDI | ||||||||
| CYP2C19 NM + CYP3A4 NM + Yes CYP Modulator | 7 (8.8) | 10 (7.1) | 0.98 (0.34–2.83) | 0.96 | 0.96 | 1.03 (0.34–3.12) | 0.96 | 0.96 |
| DGI | ||||||||
| CYP2C19 IM/PM + CYP3A4 NM + No CYP Modulator | 18 (22.5) | 42 (30) | 0.59 (0.29–1.22) | 0.16 | 0.61 | 0.69 (0.33–1.45) | 0.33 | 0.54 |
| CYP2C19 IM/PM + CYP3A4 IM/PM + No CYP Modulator | 6 (7.5) | 13 (9.3) | 0.64 (0.22–1.87) | 0.42 | 0.61 | 0.64 (0.21–1.91) | 0.42 | 0.55 |
| CYP2C19 NM + CYP3A4 IM/PM + No CYP Modulator | 10 (12.5) | 9 (6.4) | 1.55 (0.57–4.23) | 0.39 | 0.61 | 1.60 (0.56–4.56) | 0.37 | 0.54 |
| CYP2C19 UM + CYP3A4 NM/IM + No CYP Modulator | 2 (2.5) | 4 (2.9) | 0.69 (0.12–4.03) | 0.69 | 0.73 | 0.72 (0.12–4.35) | 0.72 | 0.82 |
| DDGI | 4 (5) | 16 (11.4) | 0.35 (0.11–1.14) | 0.08 | 0.45 | 0.48 (0.14–1.61) | 0.23 | 0.54 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bahar, M.A.; Lanting, P.; Bos, J.H.J.; Sijmons, R.H.; Hak, E.; Wilffert, B. Impact of Drug-Gene-Interaction, Drug-Drug-Interaction, and Drug-Drug-Gene-Interaction on (es)Citalopram Therapy: The PharmLines Initiative. J. Pers. Med. 2020, 10, 256. https://doi.org/10.3390/jpm10040256
Bahar MA, Lanting P, Bos JHJ, Sijmons RH, Hak E, Wilffert B. Impact of Drug-Gene-Interaction, Drug-Drug-Interaction, and Drug-Drug-Gene-Interaction on (es)Citalopram Therapy: The PharmLines Initiative. Journal of Personalized Medicine. 2020; 10(4):256. https://doi.org/10.3390/jpm10040256
Chicago/Turabian StyleBahar, Muh. Akbar, Pauline Lanting, Jens H. J. Bos, Rolf H. Sijmons, Eelko Hak, and Bob Wilffert. 2020. "Impact of Drug-Gene-Interaction, Drug-Drug-Interaction, and Drug-Drug-Gene-Interaction on (es)Citalopram Therapy: The PharmLines Initiative" Journal of Personalized Medicine 10, no. 4: 256. https://doi.org/10.3390/jpm10040256
APA StyleBahar, M. A., Lanting, P., Bos, J. H. J., Sijmons, R. H., Hak, E., & Wilffert, B. (2020). Impact of Drug-Gene-Interaction, Drug-Drug-Interaction, and Drug-Drug-Gene-Interaction on (es)Citalopram Therapy: The PharmLines Initiative. Journal of Personalized Medicine, 10(4), 256. https://doi.org/10.3390/jpm10040256