Systematic Search for Novel Circulating Biomarkers Associated with Extracellular Vesicles in Alzheimer’s Disease: Combining Literature Screening and Database Mining Approaches
Abstract
:1. Introduction
2. Methods
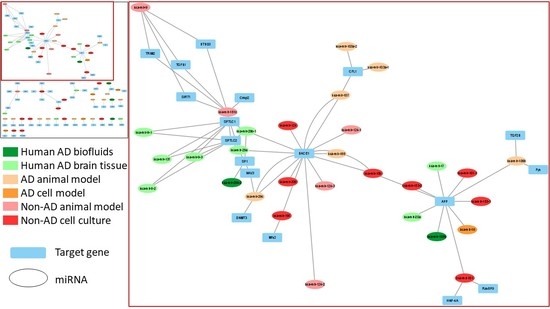
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Förstl, H.; Kurz, A. Clinical features of Alzheimer’s disease. Eur. Arch. Psychiatry Clin. Neurosci. 1999, 249, 288–290. [Google Scholar] [CrossRef]
- Mayeux, R.; Stern, Y. Epidemiology of Alzheimer Disease. Cold Spring Harb. Perspect. Med. 2012, 2, a006239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kocahan, S.; Doğan, Z. Mechanisms of Alzheimer’s Disease Pathogenesis and Prevention: The Brain, Neural Pathology, N-methyl-D-aspartate Receptors, Tau Protein and Other Risk Factors. Clin. Psychopharmacol. Neurosci. 2017, 15, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Chaudhury, S.; Brookes, K.J.; Patel, T.; Fallows, A.; Guetta-Baranes, T.; Turton, J.C.; Guerreiro, R.; Bras, J.; Hardy, J.; Francis, P.T.; et al. Alzheimer’s disease polygenic risk score as a predictor of conversion from mild-cognitive impairment. Transl. Psychiatry 2019, 9, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Jiao, B.; Liu, X.; Zhou, L.; Wang, M.H.; Zhou, Y.; Xiao, T.; Zhang, W.; Sun, R.; Waye, M.M.Y.; Tang, B.; et al. Polygenic Analysis of Late-Onset Alzheimer’s Disease from Mainland China. PLoS ONE 2015, 10, e0144898. [Google Scholar] [CrossRef] [PubMed]
- Marden, J.R.; Walter, S.; Tchetgen, E.J.T.; Kawachi, I.; Glymour, M.M. Validation of a polygenic risk score for dementia in black and white individuals. Brain Behav. 2014, 4, 687–697. [Google Scholar] [CrossRef] [Green Version]
- Lambert, J.C.; Ibrahim-Verbaas, C.A.; Harold, D.; Naj, A.C.; Sims, R.; Bellenguez, C.; DeStafano, A.L.; Bis, J.C.; Beecham, G.W.; Grenier-Boley, B.; et al. Meta-Analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 2013, 45, 1452–1458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jansen, I.E.; Savage, J.E.; Watanabe, K.; Bryois, J.; Williams, D.; Steinberg, S.; Sealock, J.; Karlsson, I.; Hägg, S.; Athanasiu, L.; et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 2019, 51, 404–413. [Google Scholar] [CrossRef] [PubMed]
- Kunkle, B.W.; Grenier-Boley, B.; Sims, R.; Bis, J.C.; Damotte, V.; Naj, A.C.; Boland, A.; Vronskaya, M.; Van Der Lee, S.J.; Amlie-Wolf, A.; et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 2019, 51, 414–430. [Google Scholar] [CrossRef] [Green Version]
- Takousis, P.; Sadlon, A.; Schulz, J.; Wohlers, I.; Dobricic, V.; Middleton, L.; Lill, C.M.; Perneczky, R.; Bertram, L. Differential expression of microRNAs in Alzheimer’s disease brain, blood, and cerebrospinal fluid. Alzheimer’s Dement. 2019, 15, 1468–1477. [Google Scholar] [CrossRef]
- Silvestro, S.; Bramanti, P.; Mazzon, E. Role of miRNAs in Alzheimer’s Disease and Possible Fields of Application. Int. J. Mol. Sci. 2019, 20, 3979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, M.; Zhang, Q.; Deng, M.; Miao, J.; Guo, Y.; Gao, W.; Cui, Q. An Analysis of Human MicroRNA and Disease Associations. PLoS ONE 2008, 3, e3420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.-Y.; Lin, Y.-C.-D.; Li, J.; Huang, K.-Y.; Shrestha, S.; Hong, H.-C.; Tang, Y.; Chen, Y.-G.; Jin, C.-N.; Yu, Y.; et al. miRTarBase 2020: Updates to the experimentally validated microRNA–target interaction database. Nucleic Acids Res. 2020, 48, D148–D154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jack, C.R., Jr.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Dunn, B.; Haeberlein, S.B.; Holtzman, D.M.; Jagust, W.; Jessen, F.; Karlawish, J.; et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimer’s Dement. 2018, 14, 535–562. [Google Scholar] [CrossRef]
- Masters, C.L.; Bateman, R.; Blennow, K.; Rowe, C.C.; Sperling, R.A.; Cummings, J.L. Alzheimer’s disease. Nat. Rev. Dis. Prim. 2015, 1, 15056. [Google Scholar] [CrossRef]
- Dubois, B.; Hampel, H.; Feldman, H.H.; Scheltens, P.; Aisen, P.; Andrieu, S.; Bakardjian, H.; Benali, H.; Bertram, L.; Blennow, K.; et al. Preclinical Alzheimer’s disease: Definition, natural history, and diagnostic criteria. Alzheimer’s Dement. J. Alzheimer’s Assoc. 2016, 12, 292–323. [Google Scholar] [CrossRef]
- Agnello, L.; Piccoli, T.; Vidali, M.; Cuffaro, L.; Sasso, B.L.; Iacolino, G.; Giglio, V.R.; Lupo, F.; Alongi, P.; Bivona, G.; et al. Diagnostic accuracy of cerebrospinal fluid biomarkers measured by chemiluminescent enzyme immunoassay for Alzheimer disease diagnosis. Scand. J. Clin. Lab. Investig. 2020, 80, 313–317. [Google Scholar] [CrossRef]
- Mattsson, N.; Andreasson, U.; Zetterberg, H.; Blennow, K. Association of Plasma Neurofilament Light With Neurodegeneration in Patients With Alzheimer Disease. JAMA Neurol. 2017, 74, 557–566. [Google Scholar] [CrossRef]
- Lewczuk, P.; Matzen, A.; Blennow, K.; Parnetti, L.; Molinuevo, J.L.; Eusebi, P.; Kornhuber, J.; Morris, J.C.; Fagan, A.M. Cere-brospinal Fluid A 42/40 Corresponds Better than A 42 to Amyloid PET in Alzheimer’s Disease the Creative Commons Attrib-ution Non-Commercial License (CC BY-NC 4.0). J. Alzheimer’s Dis. 2017, 55, 813–822. [Google Scholar] [CrossRef] [Green Version]
- Agnello, L.; Gambino, C.M.; Sasso, B.L.; Bivona, G.; Milano, S.; Ciaccio, A.M.; Piccoli, T.; La Bella, V.; Ciaccio, M. Neurogranin as a Novel Biomarker in Alzheimer’s Disease. Lab. Med. 2021, 52, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Agnello, L.; Colletti, T.; Sasso, B.L.; Vidali, M.; Spataro, R.; Gambino, C.M.; Giglio, R.V.; Piccoli, T.; Bivona, G.; La Bella, V.; et al. Tau protein as a diagnostic and prognostic biomarker in amyotrophic lateral sclerosis. Eur. J. Neurol. 2021, 28, 1868–1875. [Google Scholar] [CrossRef] [PubMed]
- Vieira, D.; Durães, J.; Baldeiras, I.; Santiago, B.; Duro, D.; Lima, M.; Leitão, M.J.; Tábuas-Pereira, M.; Santana, I. Lower CSF Amyloid-Beta1–42 Predicts a Higher Mortality Rate in Frontotemporal Dementia. Diagnostics 2019, 9, 162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Virgilio, E.; Vecchio, D.; Crespi, I.; Serino, R.; Cantello, R.; Dianzani, U.; Comi, C. Cerebrospinal Tau levels as a predictor of early disability in multiple sclerosis. Mult. Scler. Relat. Disord. 2021, 56, 103231. [Google Scholar] [CrossRef] [PubMed]
- Denk, J.; Boelmans, K.; Siegismund, C.S.; Lassner, D.; Arlt, S.; Jahn, H. MicroRNA Profiling of CSF Reveals Potential Biomarkers to Detect Alzheimer’s Disease. PLoS ONE 2015, 10, e0126423. [Google Scholar] [CrossRef] [Green Version]
- Graykowski, D.R.; Wang, Y.-Z.; Upadhyay, A.; Savas, J.N. The Dichotomous Role of Extracellular Vesicles in the Central Nervous System. iScience 2020, 23, 101456. [Google Scholar] [CrossRef]
- Rajendran, L.; Honsho, M.; Zahn, T.R.; Keller, P.; Geiger, K.D.; Verkade, P.; Simons, K. Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc. Natl. Acad. Sci. USA 2006, 103, 11172–11177. [Google Scholar] [CrossRef] [Green Version]
- An, K.; Klyubin, I.; Kim, Y.; Jung, J.H.; Mably, A.J.; O’Dowd, S.T.; Lynch, T.; Kanmert, D.; A Lemere, C.; Finan, G.M.; et al. Exosomes neutralize synaptic-plasticity-disrupting activity of Aβ assemblies in vivo. Mol. Brain 2013, 6, 47. [Google Scholar] [CrossRef] [Green Version]
- Dinkins, M.B.; Dasgupta, S.; Wang, G.; Zhu, G.; Bieberich, E. Exosome reduction in vivo is associated with lower amyloid plaque load in the 5XFAD mouse model of Alzheimer’s disease. Neurobiol. Aging 2014, 35, 1792–1800. [Google Scholar] [CrossRef] [Green Version]
- Saman, S.; Kim, W.; Raya, M.; Visnick, Y.; Miro, S.; Saman, S.; Jackson, B.; McKee, A.C.; Alvarez, V.E.; Lee, N.C.; et al. Exosome-associated Tau Is Secreted in Tauopathy Models and Is Selectively Phosphorylated in Cerebrospinal Fluid in Early Alzheimer Disease. J. Biol. Chem. 2012, 287, 3842–3849. [Google Scholar] [CrossRef] [Green Version]
- Badhwar, A.; Haqqani, A.S. Biomarker potential of brain-secreted extracellular vesicles in blood in Alzheimer’s disease. Alzheimer’s Dementia Diagn. Assess. Dis. Monit. 2020, 12, e12001. [Google Scholar] [CrossRef]
- D’Anca, M.; Fenoglio, C.; Serpente, M.; Arosio, B.; Cesari, M.; Scarpini, E.A.; Galimberti, D. Exosome Determinants of Physiological Aging and Age-Related Neurodegenerative Diseases. Front. Aging Neurosci. 2019, 11, 232. [Google Scholar] [CrossRef] [Green Version]
- Müller, M.; Kuiperij, H.B.; Versleijen, A.A.; Chiasserini, D.; Farotti, L.; Baschieri, F.; Parnetti, L.; Struyfs, H.; De Roeck, N.; Luyckx, J.; et al. Validation of microRNAs in Cerebrospinal Fluid as Biomarkers for Different Forms of Dementia in a Multicenter Study. J. Alzheimer’s Dis. 2016, 52, 1321–1333. [Google Scholar] [CrossRef]
- Cao, F.; Liu, Z.; Sun, G. Diagnostic value of miR-193a-3p in Alzheimer’s disease and miR-193a-3p attenuates amyloid-β induced neurotoxicity by targeting PTEN. Exp. Gerontol. 2020, 130, 110814. [Google Scholar] [CrossRef] [PubMed]
- Riancho, J.; Vázquez-Higuera, J.L.; Pozueta, A.; Lage, C.; Kazimierczak, M.; Bravo, M.; Calero, M.; Gonalezález, A.; Rodríguez, E.; Lleó, A.; et al. MicroRNA Profile in Patients with Alzheimer’s Disease: Analysis of miR-9-5p and miR-598 in Raw and Exosome Enriched Cerebrospinal Fluid Samples. J. Alzheimer’s Dis. 2017, 57, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Derkow, K.; Rössling, R.; Schipke, C.; Krüger, C.; Bauer, J.; Fähling, M.; Stroux, A.; Schott, E.; Ruprecht, K.; Peters, O.; et al. Distinct expression of the neurotoxic microRNA family let-7 in the cerebrospinal fluid of patients with Alzheimer’s disease. PLoS ONE 2018, 13, e0200602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogrinc, D.; Goričar, K.; Dolžan, V. Genetic Variability in Molecular Pathways Implicated in Alzheimer’s Disease: A Comprehensive Review. Front. Aging Neurosci. 2021, 13, 13. [Google Scholar] [CrossRef] [PubMed]
- Oliveros, J.C. Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 4 May 2021).
- Tsukamoto, Y.; Nakada, C.; Noguchi, T.; Tanigawa, M.; Nguyen, L.T.; Uchida, T.; Hijiya, N.; Matsuura, K.; Fujioka, T.; Seto, M.; et al. MicroRNA-375 Is Downregulated in Gastric Carcinomas and Regulates Cell Survival by Targeting PDK1 and 14-3-3ζ. Cancer Res. 2010, 70, 2339–2349. [Google Scholar] [CrossRef] [Green Version]
- Kishore, S.; Jaskiewicz, L.; Burger, L.; Hausser, J.; Khorshid, M.; Zavolan, M. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat. Methods 2011, 8, 559–564. [Google Scholar] [CrossRef]
- Sarma, N.J.; Tiriveedhi, V.; Crippin, J.S.; Chapman, W.C.; Mohanakumar, T. Hepatitis C Virus-Induced Changes in MicroRNA 107 (miRNA-107) and miRNA-449a Modulate CCL2 by Targeting the Interleukin-6 Receptor Complex in Hepatitis. J. Virol. 2014, 88, 3733–3743. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.-G.; Song, J.; Zhang, Y.-Q.; Wang, P.-C. MicroRNA-193b is a regulator of amyloid precursor protein in the blood and cerebrospinal fluid derived exosomal microRNA-193b is a biomarker of Alzheimer’s disease. Mol. Med. Rep. 2014, 10, 2395–2400. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Song, Y.; Zhou, X.; Deng, Y.; Liu, T.; Weng, G.; Yu, D.; Pan, S. MicroRNA-29c targets β-site amyloid precursor protein-cleaving enzyme 1 and has a neuroprotective role in vitro and in vivo. Mol. Med. Rep. 2015, 12, 3081–3088. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Li, X.; Xin, X.; Kan, P.-C.; Yan, Y. MicroRNA-613 regulates the expression of brain-derived neurotrophic factor in Alzheimer’s disease. Biosci. Trends 2016, 10, 372–377. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Song, Y.; Zhou, X.; Deng, Y.; Liu, T.; Weng, G.; Yu, D.; Pan, S. DNA methyltransferase 3, a target of microRNA-29c, contributes to neuronal proliferation by regulating the expression of brain-derived neurotrophic factor. Mol. Med. Rep. 2015, 12, 1435–1442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, H.; Guo, S.; Zhang, Y.; Zheng, Z.; Wang, H. Upregulation of microRNA-206 enhances lipopolysaccharide-induced inflammation and release of amyloid-β by targeting insulin-like growth factor 1 in microglia. Mol. Med. Rep. 2016, 14, 1357–1364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geng, L.; Zhang, T.; Liu, W.; Chen, Y. Inhibition of miR-128 Abates Aβ-Mediated Cytotoxicity by Targeting PPAR-γ via NF-κB Inactivation in Primary Mouse Cortical Neurons and Neuro2a Cells. Yonsei Med. J. 2018, 59, 1096–1106. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, A.; Xia, C.; Lin, Q.; Chen, C. A single nucleotide polymorphism in primary-microRNA-146a reduces the expression of mature microRNA-146a in patients with Alzheimer’s disease and is associated with the pathogenesis of Alzheimer’s disease. Mol. Med. Rep. 2015, 12, 4037–4042. [Google Scholar] [CrossRef] [Green Version]
- Dehury, B.; Kepp, K.P. Membrane dynamics of γ-secretase with the anterior pharynx-defective 1B subunit. J. Cell. Biochem. 2021, 122, 69–85. [Google Scholar] [CrossRef]
- Acx, H.; Serneels, L.; Radaelli, E.; Muyldermans, S.; Vincke, C.; Pepermans, E.; Müller, U.; Chávez-Gutiérrez, L.; De Strooper, B. Inactivation of γ-secretases leads to accumulation of substrates and non-Alzheimer neurodegeneration. EMBO Mol. Med. 2017, 9, 1088–1099. [Google Scholar] [CrossRef]
- Schwartzentruber, J.; Cooper, S.; Liu, J.Z.; Barrio-Hernandez, I.; Bello, E.; Kumasaka, N.; Young, A.M.H.; Franklin, R.J.M.; Johnson, T.; Estrada, K.; et al. Genome-wide meta-analysis, fine-mapping and integrative prioritization implicate new Alzheimer’s disease risk genes. Nat. Genet. 2021, 53, 392–402. [Google Scholar] [CrossRef]
- Yanaizu, M.; Washizu, C.; Nukina, N.; Satoh, J.-I.; Kino, Y. CELF2 regulates the species-specific alternative splicing of TREM. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Ulland, T.K.; Colonna, M. TREM2—A key player in microglial biology and Alzheimer disease. Nat. Rev. Neurol. 2018, 14, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, T.; Stefansson, H.; Steinberg, S.; Jonsdottir, I.; Jonsson, P.V.; Snaedal, J.; Bjornsson, S.; Huttenlocher, J.; Levey, A.I.; Lah, J.J.; et al. Variant of TREM2 Associated with the Risk of Alzheimer’s Disease. N. Engl. J. Med. 2013, 368, 107–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sims, R.; Van Der Lee, S.J.; Naj, A.C.; Bellenguez, C.; Badarinarayan, N.; Jakobsdottir, J.; Kunkle, B.W.; Boland, A.; Raybould, R.; Bis, J.C.; et al. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat. Genet. 2017, 49, 1373–1384. [Google Scholar] [CrossRef] [PubMed]
- Wijsman, E.M.; Pankratz, N.; Choi, Y.; Rothstein, J.H.; Faber, K.M.; Cheng, R.; Lee, J.H.; Bird, T.D.; Bennett, D.A.; Diaz-Arrastia, R.; et al. Genome-Wide Association of Familial Late-Onset Alzheimer’s Disease Replicates BIN1 and CLU and Nominates CUGBP2 in Interaction with APOE. PLoS Genet. 2011, 7, e1001308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Ge, X.; Zhang, J.; Chen, L. Effect of lncRNA WT1-AS regulating WT1 on oxidative stress injury and apoptosis of neurons in Alzheimer’s disease via inhibition of the miR-375/SIX4 axis. Aging 2020, 12, 23974–23995. [Google Scholar] [CrossRef]
- Pirone, D.M.; Fukuhara, S.; Gutkind, J.S.; Burbelo, P.D. SPECs, Small Binding Proteins for Cdc. J. Biol. Chem. 2000, 275, 22650–22656. [Google Scholar] [CrossRef] [Green Version]
- Bamburg, J.R.; Bloom, G.S. Cytoskeletal pathologies of Alzheimer disease. Cell Motil. Cytoskelet. 2009, 66, 635–649. [Google Scholar] [CrossRef] [Green Version]
- Rush, T.; Martinez-Hernandez, J.; Dollmeyer, M.; Frandemiche, M.L.; Borel, E.; Boisseau, S.; Jacquier-Sarlin, M.; Buisson, A. Synaptotoxicity in Alzheimer’s Disease Involved a Dysregulation of Actin Cytoskeleton Dynamics through Cofilin 1 Phosphorylation. J. Neurosci. 2018, 38, 10349–10361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jun, G.; Ibrahim-Verbaas, C.A.; Vronskaya, M.; Lambert, J.C.; Chung, J.; Naj, A.C.; Kunkle, B.W.; Wang, L.S.; Bis, J.C.; Bel-lenguez, C.; et al. A novel Alzheimer disease locus located near the gene encoding tau protein. Mol. Psychiatry 2016, 21, 108–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, J.A.; Wood, P.L.; Ryan, R.; Graff-Radford, N.R.; Pilapil, C.; Robitaille, Y.; Quirion, R. Cytokine indices in Alzheimer’s temporal cortex: No changes in mature IL-1β or IL-1RA but increases in the associated acute phase proteins IL-6, α2-macroglobulin and C-reactive protein. Brain Res. 1993, 629, 245–252. [Google Scholar] [CrossRef]
- Hull, M.; Berger, M.; Volk, B.; Bauer, J. Occurrence of Interleukin-6 in Cortical Plaques of Alzheimer’s Disease Patients May Precede Transformation of Diffuse into Neuritic Plaquesa. Ann. N. Y. Acad. Sci. 1996, 777, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Islam, O.; Gong, X.; Rose-John, S.; Heese, K. Interleukin-6 and Neural Stem Cells: More Than Gliogenesis. Mol. Biol. Cell 2009, 20, 188–199. [Google Scholar] [CrossRef]
- Yang, R.; Duan, J.; Luo, F.; Tao, P.; Hu, C. IL-6, IL-8 and IL-10 polymorphisms may impact predisposition of Alzheimer’s disease: A meta-analysis. Acta Neurol. Belg. 2020, 1–8. [Google Scholar] [CrossRef]
- Kauwe, J.S.K.; Bailey, M.H.; Ridge, P.G.; Perry, R.; Wadsworth, M.E.; Hoyt, K.L.; Staley, L.A.; Karch, C.; Harari, O.; Cruchaga, C.; et al. Genome-Wide Association Study of CSF Levels of 59 Alzheimer’s Disease Candidate Proteins: Significant Associations with Proteins Involved in Amyloid Processing and Inflammation. PLoS Genet. 2014, 10, e1004758. [Google Scholar] [CrossRef]
- Haddick, P.C.; Larson, J.L.; Rathore, N.; Bhangale, T.R.; Phung, Q.T.; Srinivasan, K.; Hansen, D.V.; Lill, J.R.; Pericak-Vance, M.A.; Haines, J.; et al. A Common Variant of IL-6R is Associated with Elevated IL-6 Pathway Activity in Alzheimer’s Disease Brains. J. Alzheimer’s Dis. 2017, 56, 1037–1054. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Wu, L.; Hu, Y.; Jiang, L.; Liang, N.; Chen, J.; Qin, H.; Tang, N. MicroRNA-107 Ameliorates Damage in a Cell Model of Alzheimer’s Disease by Mediating the FGF7/FGFR2/PI3K/Akt Pathway. J. Mol. Neurosci. 2020, 70, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, C.; Zhang, Y. An investigation of microRNA-103 and microRNA-107 as potential blood-based biomarkers for disease risk and progression of Alzheimer’s disease. J. Clin. Lab. Anal. 2020, 34, e23006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, B.; Zhou, H.; Zhang, R.; Song, M.; Yu, L.; Wang, L.; Liu, Z.; Zhang, Q.; Cui, D.; Wang, X.; et al. Serum miR-206 and miR-132 as Potential Circulating Biomarkers for Mild Cognitive Impairment. J. Alzheimer’s Dis. 2015, 45, 721–731. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.T.; Liu, C.G.; Gao, S.C.; Zhang, Y.; Wang, P.C. The Serum Exosome Derived MicroRNA-135a, -193b, and -384 Were Potential Alzheimer’s Disease Biomarkers. Biomed. Environ. Sci. 2018, 31, 87–96. [Google Scholar] [PubMed]
- Montagna, E.; Dorostkar, M.M.; Herms, J. The Role of APP in Structural Spine Plasticity. Front. Mol. Neurosci. 2017, 10, 136. [Google Scholar] [CrossRef] [Green Version]
- Haass, C. Take five—BACE and the γ-secretase quartet conduct Alzheimer’s amyloid β-peptide generation. EMBO J. 2004, 23, 483–488. [Google Scholar] [CrossRef]
- Ewers, M.; Cheng, X.; Zhong, Z.; Nural, H.F.; Walsh, C.; Meindl, T.; Teipel, S.J.; Buerger, K.; He, P.; Shen, Y.; et al. Increased CSF-BACE1 Activity Associated with Decreased Hippocampus Volume in Alzheimer’s Disease. J. Alzheimer’s Dis. 2011, 25, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Ewers, M.; Zhong, Z.; Bürger, K.; Wallin, A.; Blennow, K.; Teipel, S.J.; Shen, Y.; Hampel, H. Increased CSF-BACE 1 activity is associated with ApoE-ε4 genotype in subjects with mild cognitive impairment and Alzheimer’s disease. Brain 2008, 131, 1252–1258. [Google Scholar] [CrossRef] [Green Version]
- Alexopoulos, P.; Thierjung, N.; Grimmer, T.; Ortner, M.; Economou, P.; Assimakopoulos, K.; Gourzis, P.; Politis, A.; Perneczky, R.; Initiative, T.A.D.N. Cerebrospinal Fluid BACE1 Activity and sAβPPβ as Biomarker Candidates of Alzheimer’s Disease. Dement. Geriatr. Cogn. Disord. 2018, 45, 152–161. [Google Scholar] [CrossRef]
- Zhong, Z.; Ewers, M.; Teipel, S.; Bürger, K.; Wallin, A.; Blennow, K.; He, P.; McAllister, C.; Hampel, H.; Shen, Y. Levels of β-Secretase (BACE1) in Cerebrospinal Fluid as a Predictor of Risk in Mild Cognitive Impairment. Arch. Gen. Psychiatry 2007, 64, 718–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, G.; Sankaranarayanan, S.; Wong, J.; Tugusheva, K.; Michener, M.S.; Shi, X.; Cook, J.J.; Simon, A.J.; Savage, M.J. Characterization of plasma β-secretase (BACE1) activity and soluble amyloid precursor proteins as potential biomarkers for Alzheimer’s disease. J. Neurosci. Res. 2012, 90, 2247–2258. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Liu, Y.; Shen, J.; Lv, D.; Zhang, J. Meta-analysis of BACE1 gene rs638405 polymorphism and the risk of Alzheimer’s disease in Caucasion and Asian population. Neurosci. Lett. 2016, 616, 189–196. [Google Scholar] [CrossRef]
- Maxwell, T.J.; Initiative, A.D.N.; Corcoran, C.; Del-Aguila, J.L.; Budde, J.P.; Deming, Y.; Cruchaga, C.; Goate, A.M.; Kauwe, J.S.K. Genome-wide association study for variants that modulate relationships between cerebrospinal fluid amyloid-beta 42, tau, and p-tau levels. Alzheimer’s Res. Ther. 2018, 10, 86. [Google Scholar] [CrossRef]
- Saravanaraman, P.; Selvam, M.; Ashok, C.; Srijyothi, L.; Baluchamy, S. De novo methyltransferases: Potential players in diseases and new directions for targeted therapy. Biochimie 2020, 176, 85–102. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Xu, J.; Xu, J.; Cheng, J.; Jiao, D.; Zhou, C.; Dai, Y.; Chen, Q. Lower Serum Levels of miR-29c-3p and miR-19b-3p as Biomarkers for Alzheimer’s Disease. Tohoku J. Exp. Med. 2017, 242, 129–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cattaneo, A.; Cattane, N.; Begni, V.; Pariante, C.M.; A Riva, M. The human BDNF gene: Peripheral gene expression and protein levels as biomarkers for psychiatric disorders. Transl. Psychiatry 2016, 6, e958. [Google Scholar] [CrossRef]
- Hock, C.; Heese, K.; Hulette, C.; Rosenberg, C.; Otten, U. Region-Specific Neurotrophin Imbalances in Alzheimer Disease. Arch. Neurol. 2000, 57, 846–851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miranda, M.; Morici, J.F.; Zanoni, M.B.; Bekinschtein, P. Brain-Derived Neurotrophic Factor: A Key Molecule for Memory in the Healthy and the Pathological Brain. Front. Cell. Neurosci. 2019, 13, 363. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, S.; Arai, H.; Matsui, T.; Yuzuriha, T.; Urakami, K.; Masaki, T.; Higuchi, S. Brain-derived neurotrophic factor gene polymorphisms and Alzheimer’s disease. J. Neural Transm. 2005, 112, 703–771. [Google Scholar] [CrossRef] [PubMed]
- Voineskos, A.N.; Lerch, J.P.; Felsky, D.; Shaikh, S.; Rajji, T.K.; Miranda, D.; Lobaugh, N.J.; Mulsant, B.H.; Pollock, B.G.; Kennedy, J.L. The Brain-Derived Neurotrophic Factor Val66Met Polymorphism and Prediction of Neural Risk for Alzheimer Disease. Arch. Gen. Psychiatry 2011, 68, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.S.; Cai, D. Integrated miRNA-Seq and mRNA-Seq Study to Identify miRNAs Associated With Alzheimer’s Disease Using Post-mortem Brain Tissue Samples. Front. Neurosci. 2021, 15, 1–14. [Google Scholar] [CrossRef]
- Liang, C.; Zou, T.; Zhang, M.; Fan, W.; Zhang, T.; Jiang, Y.; Cai, Y.; Chen, F.; Chen, X.; Sun, Y.; et al. MicroRNA-146a switches microglial phenotypes to resist the pathological processes and cognitive degradation of Alzheimer’s disease. Theranostics 2021, 11, 4103–4121. [Google Scholar] [CrossRef]
- Cui, L.; Li, Y.; Ma, G.; Wang, Y.; Cai, Y.; Liu, S.; Chen, Y.; Li, J.; Xie, Y.; Liu, G.; et al. A Functional Polymorphism in the Promoter Region of MicroRNA-146a Is Associated with the Risk of Alzheimer Disease and the Rate of Cognitive Decline in Patients. PLoS ONE 2014, 9, e89019. [Google Scholar] [CrossRef]
- Khalilzadeh, B.; Rashidi, M.; Soleimanian, A.; Tajalli, H.; Kanberoglu, G.S.; Baradaran, B.; Rashidi, M.R. Development of a reliable microRNA based electrochemical genosensor for monitoring of miR-146a, as key regulatory agent of neurodegenerative disease. Int. J. Biol. Macromol. 2019, 134, 695–703. [Google Scholar] [CrossRef]
- Ansari, A.; Maffioletti, E.; Milanesi, E.; Marizzoni, M.; Frisoni, G.B.; Blin, O.; Richardson, J.C.; Bordet, R.; Forloni, G.; Gennarelli, M.; et al. miR-146a and miR-181a are involved in the progression of mild cognitive impairment to Alzheimer’s disease. Neurobiol. Aging 2019, 82, 102–109. [Google Scholar] [CrossRef]
- De Oliviera Nascimento, L.; Massari, P.; Wetzler, L.M. The Role of TLR2 in Infection and Immunity. Front. Immunol. 2012, 3, 79. [Google Scholar] [CrossRef] [Green Version]
- Pourbadie, H.G.; Sayyah, M.; Khoshkholgh-Sima, B.; Choopani, S.; Nategh, M.; Motamedi, F.; Shokrgozar, M.A. Early minor stimulation of microglial TLR2 and TLR4 receptors attenuates Alzheimer’s disease–related cognitive deficit in rats: Behavioral, molecular, and electrophysiological evidence. Neurobiol. Aging 2018, 70, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Lax, N.; Fainstein, N.; Nishri, Y.; Ben-Zvi, A.; Ben-Hur, T. Systemic microbial TLR2 agonists induce neurodegeneration in Alzheimer’s disease mice. J. Neuroinflammation 2020, 17, 1–12. [Google Scholar] [CrossRef]
- Yu, J.-T.; Mou, S.-M.; Wang, L.-Z.; Mao, C.-X.; Tan, L. Toll-like receptor 2 -196 to -174 del polymorphism influences the susceptibility of Han Chinese people to Alzheimer’s disease. J. Neuroinflammation 2011, 8, 136. [Google Scholar] [CrossRef] [Green Version]
- Sohrabifar, N.; Gharesouran, J.; Talebi, M.; Ghojazadeh, M.; Ardebili, S.M.M. Association of CLU and TLR2 gene polymorphisms with late-onsetAlzheimer disease in a northwestern Iranian population. Turk. J. Med. Sci. 2015, 45, 1082–1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, R.; Jow, L.; Croston, G.E.; Paterniti, J.R.J. Identification, Characterization, and Tissue Distribution of Human Peroxisome Proliferator-activated Receptor (PPAR) Isoforms PPARγ2 versus PPARγ1 and Activation with Retinoid X Receptor Agonists and Antagonists. J. Biol. Chem. 1997, 272, 8071–8076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khavinson, V.; Linkova, N.; Kozhevnikova, E.; Trofimova, S. EDR Peptide: Possible Mechanism of Gene Expression and Protein Synthesis Regulation Involved in the Pathogenesis of Alzheimer’s Disease. Molecules 2020, 26, 159. [Google Scholar] [CrossRef]
- Wang, S.; Guan, L.; Luo, D.; Liu, J.; Lin, H.; Li, X.; Liu, X. Gene- gene interaction between PPARG and APOE gene on late-onset Alzheimer’s disease: A case- control study in Chinese han population. J. Nutr. Health Aging 2016, 21, 397–403. [Google Scholar] [CrossRef]
- Koivisto, A.M.; Helisalmi, S.; Pihlajamäki, J.; Hiltunen, M.; Koivisto, K.; Moilanen, L.; Kuusisto, J.; Helkala, E.-L.; Hänninen, T.; Kervinen, K.; et al. Association Analysis of Peroxisome Proliferator-Activated Receptor Gamma Polymorphisms and Late Onset Alzheimer’s Disease in the Finnish Population. Dement. Geriatr. Cogn. Disord. 2006, 22, 449–453. [Google Scholar] [CrossRef]
- Tiribuzi, R.; Crispoltoni, L.; Porcellati, S.; Di Lullo, M.; Florenzano, F.; Pirro, M.; Bagaglia, F.; Kawarai, T.; Zampolini, M.; Orlacchio, A.; et al. miR128 up-regulation correlates with impaired amyloid β(1-42) degradation in monocytes from patients with sporadic Alzheimer’s disease. Neurobiol. Aging 2014, 35, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, Y.; Liu, P.; Bai, H.; Li, X.; Xiao, J.; Yuan, Q.; Geng, S.; Yin, H.; Zhang, H.; et al. MicroRNA-128 knockout inhibits the development of Alzheimer’s disease by targeting PPARγ in mouse models. Eur. J. Pharmacol. 2019, 843, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Han, W.; Xu, Y.; Li, D.; Xue, Q. Serum miR-128 Serves as a Potential Diagnostic Biomarker for Alzheimer’s Disease. Neuropsychiatr. Dis. Treat. 2021, 17, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.-G.; Son, H. Adult hippocampal neurogenesis and related neurotrophic factors. BMB Rep. 2009, 42, 239–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westwood, A.J.; Beiser, A.; DeCarli, C.; Harris, T.B.; Chen, T.C.; He, X.-M.; Roubenoff, R.; Pikula, A.; Au, R.; Braverman, L.E.; et al. Insulin-like growth factor-1 and risk of Alzheimer dementia and brain atrophy. Neurology 2014, 82, 1613–1619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rui-Hua, C.; Yong-De, P.; Xiao-Zhen, J.; Chen, J.; Bin, Z. Decreased Levels of Serum IGF-1 and Vitamin D Are Associated With Cognitive Impairment in Patients With Type 2 Diabetes. Am. J. Alzheimer’s Dis. Other Dementiasr. 2019, 34, 450–456. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Yu, J.-T.; Tan, L.; Liu, Q.-Y.; Wang, H.-F.; Ma, X.-Y. Insulin-like growth factor 1 (IGF1) polymorphism is associated with Alzheimer’s disease in Han Chinese. Neurosci. Lett. 2012, 531, 20–23. [Google Scholar] [CrossRef] [PubMed]
- Xie, B.; Liu, Z.; Jiang, L.; Liu, W.; Song, M.; Zhang, Q.; Zhang, R.; Cui, D.; Wang, X.; Xu, S. Increased Serum miR-206 Level Predicts Conversion from Amnestic Mild Cognitive Impairment to Alzheimer’s Disease: A 5-Year Follow-up Study. J. Alzheimer’s Dis. 2016, 55, 509–520. [Google Scholar] [CrossRef]
- Moon, J.; Lee, S.-T.; Kong, I.G.; Byun, J.-I.; Sunwoo, J.-S.; Shin, J.-W.; Shim, J.-Y.; Park, J.-H.; Jeon, D.; Jung, K.-H.; et al. Early diagnosis of Alzheimer’s disease from elevated olfactory mucosal miR-206 level. Sci. Rep. 2016, 6, 20364. [Google Scholar] [CrossRef]
- Wang, C.-N.; Wang, Y.-J.; Wang, H.; Song, L.; Chen, Y.; Wang, J.-L.; Ye, Y.; Jiang, B. The Anti-dementia Effects of Donepezil Involve miR-206-3p in the Hippocampus and Cortex. Biol. Pharm. Bull. 2017, 40, 465–472. [Google Scholar] [CrossRef] [Green Version]
- Dangla-Valls, A.; Molinuevo, J.L.; Altirriba, J.; Sanchez-Valle, R.; Alcolea, D.; Fortea, J.; Rami, L.; Balasa, M.; Muñoz-García, C.; Ezquerra, M.; et al. CSF microRNA Profiling in Alzheimer’s Disease: A Screening and Validation Study. Mol. Neurobiol. 2016, 54, 6647–6654. [Google Scholar] [CrossRef] [PubMed]
- Cosín-Tomàs, M.; Antonell, A.; Lladó, A.; Alcolea, D.; Fortea, J.; Ezquerra, M.; Lleó, A.; Martí, M.J.; Pallàs, M.; Sanchez-Valle, R.; et al. Plasma miR-34a-5p and miR-545-3p as Early Biomarkers of Alzheimer’s Disease: Potential and Limitations. Mol. Neurobiol. 2017, 54, 5550–5562. [Google Scholar] [CrossRef]
- Müller, M.; Jäkel, L.; Bruinsma, I.B.; Claassen, J.A.; Kuiperij, B.; Verbeek, M.M. MicroRNA-29a Is a Candidate Biomarker for Alzheimer’s Disease in Cell-Free Cerebrospinal Fluid. Mol. Neurobiol. 2016, 53, 2894–2899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leidinger, P.; Backes, C.; Deutscher, S.; Schmitt, K.; Mueller, S.C.; Frese, K.; Haas, J.; Ruprecht, K.; Paul, F.; Stähler, C.; et al. A blood based 12-miRNA signature of Alzheimer disease patients. Genome Biol. 2013, 14, R78. [Google Scholar] [CrossRef] [Green Version]
- Denk, J.; Oberhauser, F.; Kornhuber, J.; Wiltfang, J.; Fassbender, K.; Schroeter, M.L.; Volk, A.E.; Diehl-Schmid, J.; Prudlo, J.; Danek, A.; et al. Specific serum and CSF microRNA profiles distinguish sporadic behavioural variant of frontotemporal de-mentia compared with Alzheimer patients and cognitively healthy controls. PLoS ONE 2018, 13, e0197329. [Google Scholar] [CrossRef] [PubMed]
- Manzine, P.R.; Pelucchi, S.; Horst, M.A.; Vale, F.A.; Pavarini, S.C.; Audano, M.; Mitro, N.; Di Luca, M.; Marcello, E.; Cominetti, M.R. microRNA 221 Targets ADAM10 mRNA and is Downregulated in Alzheimer’s Disease. J. Alzheimer’s Dis. 2017, 61, 113–123. [Google Scholar] [CrossRef]
- Gui, Y.; Liu, H.; Zhang, L.; Lv, W.; Hu, X. Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget 2015, 6, 37043–37053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Q.; Zhao, Q.; Yin, Y. miR-133b is a potential diagnostic biomarker for Alzheimer’s disease and has a neuroprotective role. Exp. Ther. Med. 2019, 18, 2711–2718. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| miRNA | Target Gene | Method | PubMed ID (Reference) |
|---|---|---|---|
| hsa-miR-375 | APH1B | Microarray | 20215506 [38] |
| hsa-miR-375 | CELF2 | Microarray | 20215506 [38] |
| hsa-miR-107 | CDC42SE2 | PAR-CLIP 1 | 21572407 [39] |
| hsa-miR-107 | IL6 | Luciferase reporter assay, RT-qPCR 2, Western blot | 24429361 [40] |
| miRNA | Target Gene | Method | PubMed ID (Reference) |
|---|---|---|---|
| hsa-miR-193b | APP | Luciferase reporter assay, RT-qPCR 1, Western blot | 25119742 [41] |
| hsa-miR-29c | BACE1 | Luciferase reporter assay, RT-qPCR 1, Western blot | 25955795 [42] |
| hsa-miR-613 | BDNF | EGFP reporter assay, RT-qPCR 1, Western blot | 27545218 [43] |
| hsa-miR-29c | DNMT3 | Luciferase reporter assay, RT-qPCR 1, Western blot | 25815896 [44] |
| hsa-miR-206 | IGF1 | Luciferase reporter assay, RT-qPCR 1, Western blot | 27277332 [45] |
| hsa-miR-128 | PPARG | Luciferase reporter assay, RT-qPCR 1, Western blot | 30328325 [46] |
| hsa-miR-146a | TLR2 | Luciferase reporter assay, RT-qPCR 1, Western blot | 26095531 [47] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vogrinc, D.; Goričar, K.; Kunej, T.; Dolžan, V. Systematic Search for Novel Circulating Biomarkers Associated with Extracellular Vesicles in Alzheimer’s Disease: Combining Literature Screening and Database Mining Approaches. J. Pers. Med. 2021, 11, 946. https://doi.org/10.3390/jpm11100946
Vogrinc D, Goričar K, Kunej T, Dolžan V. Systematic Search for Novel Circulating Biomarkers Associated with Extracellular Vesicles in Alzheimer’s Disease: Combining Literature Screening and Database Mining Approaches. Journal of Personalized Medicine. 2021; 11(10):946. https://doi.org/10.3390/jpm11100946
Chicago/Turabian StyleVogrinc, David, Katja Goričar, Tanja Kunej, and Vita Dolžan. 2021. "Systematic Search for Novel Circulating Biomarkers Associated with Extracellular Vesicles in Alzheimer’s Disease: Combining Literature Screening and Database Mining Approaches" Journal of Personalized Medicine 11, no. 10: 946. https://doi.org/10.3390/jpm11100946
APA StyleVogrinc, D., Goričar, K., Kunej, T., & Dolžan, V. (2021). Systematic Search for Novel Circulating Biomarkers Associated with Extracellular Vesicles in Alzheimer’s Disease: Combining Literature Screening and Database Mining Approaches. Journal of Personalized Medicine, 11(10), 946. https://doi.org/10.3390/jpm11100946