Deep Learning to Detect Triangular Fibrocartilage Complex Injury in Wrist MRI: Retrospective Study with Internal and External Validation
Abstract
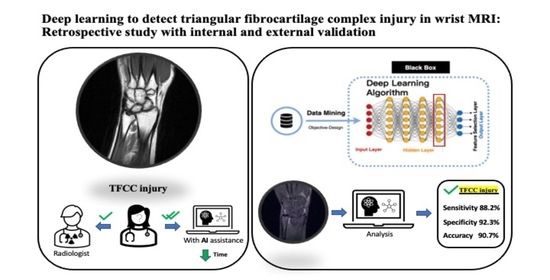
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. MRI Datasets and Radiologist Reports
2.3. Preprocessing
2.4. Algorithms and Model Architectures
2.5. Statistical Analysis
3. Results
3.1. Model Performance on the Internal and External Test Sets
3.2. Interpretation and Visualization
4. Discussion
5. Key Points
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Brahim, A.; Jennane, R.; Riad, R.; Janvier, T.; Khedher, L.; Toumi, H.; Lespessailles, E. A decision support tool for early detection of knee OsteoArthritis using X-ray imaging and machine learning: Data from the OsteoArthritis Initiative. Comput. Med. Imaging Graph. 2019, 73, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Üreten, K.; Arslan, T.; Gültekin, K.E.; Demir, A.N.D.; Özer, H.F.; Bilgili, Y. Detection of hip osteoarthritis by using plain pelvic radiographs with deep learning methods. Skeletal. Radiol. 2020, 49, 1369–1374. [Google Scholar] [CrossRef] [PubMed]
- Urakawa, T.; Tanaka, Y.; Goto, S.; Matsuzawa, H.; Watanabe, K.; Endo, N. Detecting intertrochanteric hip fractures with orthopedist-level accuracy using a deep convolutional neural network. Skeletal. Radiol. 2019, 48, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Fong, R.C.; Vedaldi, A. Interpretable explanations of black boxes by meaningful perturbation. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 3429–3437. [Google Scholar]
- Ng, A.W.H.; Griffith, J.F.; Fung, C.S.Y. MR imaging of the traumatic triangular fibrocartilaginous complex tear. Quant. Imaging Med. Surg. 2017, 7, 443–460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blazar, P.E.; Chan, P.S.; Kneeland, J.B. The effect of observer experience on magnetic resonance imaging interpretation and localization of triangular fibrocartilage complex lesions. J. Hand. Surg. Am. 2001, 26, 742–748. [Google Scholar] [CrossRef] [PubMed]
- Oren, O.; Gersh, B.J.; Bhatt, D.L. Artificial intelligence in medical imaging: Switching from radiographic pathological data to clinically meaningful endpoints. Lancet Digit. Health 2020, 2, e486–e488. [Google Scholar] [CrossRef]
- Von Schacky, C.E.; Sohn, J.; Foreman, S.C.; Liu, F.; Jungmann, P.M.; Nardo, L.; Ozhinsky, E.; Posadzy, M.; Nevitt, M.C.; Link, T.M.; et al. Development and performance comparison with radiologists of a multitask deep learning model for severity grading of hip osteoarthritis features on radiographs. Osteoarthr. Cartil. 2020, 28, S306–S308. [Google Scholar] [CrossRef]
- Ha, C.W.; Kim, S.H.; Lee, D.H.; Kim, H.; Park, Y.B. Predictive validity of radiographic signs of complete discoid lateral meniscus in children using machine learning techniques. J. Orthop. Res. 2020, 38, 1279–1288. [Google Scholar] [CrossRef]
- Namiri, N.K.; Flament, I.; Astuto, B.; Shah, R.; Tibrewala, R.; Caliva, F.; Link, T.M.; Pedoia, V.; Majumdar, S. Hierarchical severity staging of anterior cruciate ligament injuries using deep learning with MRI images. arXiv 2020, arXiv:2003.09089. [Google Scholar]
- Ambellan, F.; Tack, A.; Ehlke, M.; Zachow, S. Automated segmentation of knee bone and cartilage combining statistical shape knowledge and convolutional neural networks: Data from the Osteoarthritis Initiative. Med. Image Anal. 2019, 52, 109–118. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhao, G.; Kijowski, R.; Liu, F. Deep convolutional neural network for segmentation of knee joint anatomy. Magn. Reson. Med. 2018, 80, 2759–2770. [Google Scholar] [CrossRef] [PubMed]
- Prasoon, A.; Petersen, K.; Igel, C.; Lauze, F.; Dam, E.; Nielsen, M. Deep feature learning for knee cartilage segmentation using a triplanar convolutional neural network. Med. Image Comput. Comput-Assist. Interv. 2013, 16, 246–253. [Google Scholar] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Badgeley, M.A.; Zech, J.R.; Oakden-Rayner, L.; Glicksberg, B.S.; Liu, M.; Gale, W.; McConnell, M.V.; Percha, B.; Snyder, T.M.; Dudley, J.T. Deep learning predicts hip fracture using confounding patient and healthcare variables. NPJ Digit. Med. 2019, 2, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Q.Q.; Wang, J.; Tang, W.; Zhang-Chun, H.; Zi-Yi, X.; Xue-Song, L.; Rongguo, Z.; Xindao, Y.; Bing, Z.; Zhang, H. Automatic detection and classification of rib fractures on thoracic CT using convolutional neural network: Accuracy and feasibility. Korean J. Radiol. 2020, 21, 869–879. [Google Scholar] [CrossRef] [PubMed]
- Pedoia, V.; Norman, B.; Mehany, S.N.; Bucknor, M.D.; Link, T.M.; Majumdar, S. 3D convolutional neural networks for detection and severity staging of meniscus and PFJ cartilage morphological degenerative changes in osteoarthritis and anterior cruciate ligament subjects. J. Magn. Reson. Imaging 2019, 49, 400–410. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, M.; Zhou, Y.; Lu, G.; Zhou, Q. Deep learning approach for anterior cruciate ligament lesion detection: Evaluation of diagnostic performance using arthroscopy as the reference standard. J. Magn. Reason. Imaging 2020, 52, 1745–1752. [Google Scholar] [CrossRef]
- Weikert, T.; Noordtzij, L.A.; Bremerich, J.; Stieltjes, B.; Parmar, V.; Cyriac, J.; Sommer, G.; Sauter, A.W. Assessment of a deep learning algorithm for the detection of rib fractures on whole-body trauma computed tomography. Korean J. Radiol. 2020, 21, 891–899. [Google Scholar] [CrossRef]
- Deniz, C.M.; Xiang, S.; Hallyburton, R.S.; Welbeck, A.; Babb, J.S.; Honig, S.; Cho, K.; Chang, G. Segmentation of the proximal femur from MR images using deep convolutional neural networks. Sci. Rep. 2018, 8, 16485. [Google Scholar]
- Tiulpin, A.; Thevenot, J.; Rahtu, E.; Lehenkari, P.; Saarakkala, S. Automatic knee osteoarthritis diagnosis from plain radiographs: A deep learning-based approach. Sci. Rep. 2018, 8, 1727. [Google Scholar] [CrossRef]
- Chang, P.D.; Wong, T.T.; Rasiej, M.J. Deep learning for detection of complete anterior cruciate ligament tear. J. Digit. Imaging 2019, 32, 980–986. [Google Scholar] [CrossRef]
- Bien, N.; Rajpurkar, P.; Ball, R.; Irvin, J.; Park, A.; Jones, E.; Bereket, M.; Patel, B.N.; Yeom, K.W.; Shpanskaya, K.; et al. Deep-learning-assisted diagnosis for knee magnetic resonance imaging: Development and retrospective validation of MRNet. PloS Med. 2018, 15, e1002699. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, R.; Daluiski, A.; Chopra, S.; Lachapelle, A.; Mozer, M.; Sicular, S.; Hanel, D.; Gardner, M.; Gupta, A.; Hotchkiss, R.; et al. Deep neural network improves fracture detection by clinicians. Proc. Natl. Acad. Sci. USA 2018, 115, 11591–11596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, D.D.; Brown, E.W. Artificial intelligence in medical practice: The question to the answer? Am. J. Med. 2018, 131, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhou, Z.; Samsonov, A.; Blankenbaker, D.; Larison, W.; Kanarek, A.; Lian, K.; Kambhampati, S.; Kijowski, R. Deep learning approach for evaluating knee MR Images: Achieving high diagnostic performance for cartilage lesion detection. Radiology 2018, 289, 160–169. [Google Scholar] [CrossRef]
- Liu, F.; Zhou, Z.; Jang, H.; Samsonov, A.; Zhao, G.; Kijowski, R. Deep convolutional neural network and 3D deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging. Magn. Reson. Med. 2018, 79, 2379–2391. [Google Scholar] [CrossRef]
- Liu, F.; Guan, B.; Zhou, Z.; Samsonov, A.; Rosas, H.; Lian, K.; Sharma, R.; Kanarek, A.; Kim, J.; Guermazi, A.; et al. Fully Automated Diagnosis of Anterior Cruciate Ligament Tears on Knee MR Images by Using Deep Learning. Radiol. Artif. Intell. 2019, 1, 180091. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, K.-Y.; Li, Y.-T.; Han, J.-Y.; Wu, C.-C.; Chu, C.-M.; Peng, S.-Y.; Yeh, T.-T. Deep Learning to Detect Triangular Fibrocartilage Complex Injury in Wrist MRI: Retrospective Study with Internal and External Validation. J. Pers. Med. 2022, 12, 1029. https://doi.org/10.3390/jpm12071029
Lin K-Y, Li Y-T, Han J-Y, Wu C-C, Chu C-M, Peng S-Y, Yeh T-T. Deep Learning to Detect Triangular Fibrocartilage Complex Injury in Wrist MRI: Retrospective Study with Internal and External Validation. Journal of Personalized Medicine. 2022; 12(7):1029. https://doi.org/10.3390/jpm12071029
Chicago/Turabian StyleLin, Kun-Yi, Yuan-Ta Li, Juin-Yi Han, Chia-Chun Wu, Chi-Min Chu, Shao-Yu Peng, and Tsu-Te Yeh. 2022. "Deep Learning to Detect Triangular Fibrocartilage Complex Injury in Wrist MRI: Retrospective Study with Internal and External Validation" Journal of Personalized Medicine 12, no. 7: 1029. https://doi.org/10.3390/jpm12071029
APA StyleLin, K. -Y., Li, Y. -T., Han, J. -Y., Wu, C. -C., Chu, C. -M., Peng, S. -Y., & Yeh, T. -T. (2022). Deep Learning to Detect Triangular Fibrocartilage Complex Injury in Wrist MRI: Retrospective Study with Internal and External Validation. Journal of Personalized Medicine, 12(7), 1029. https://doi.org/10.3390/jpm12071029