Survey of Transfer Learning Approaches in the Machine Learning of Digital Health Sensing Data
Abstract
:1. Introduction
2. Digital Health Sensing Technologies
2.1. Portable Sensing Technologies
2.1.1. Wearable and Attachable Sensing Technologies
- Blood-Pressure-Monitoring (BPM) Technology
- Cardiac Monitor Technology
- Wearable Mental-Health-Monitoring Technology
- Wearable Sleep Technology
- Wearable Noninvasive Continuous-Glucose-Monitoring Technology
- Wearable Activity-Recognition Technology
- Wearable Mouth-Based Systems Technology
- Smart Shoes Technology
- Tear Biomarker Monitoring Using Eyeglasses-Nose-Bridge Pad Technology
- Attachable Patch/Bands for Sweat-Biomarker-Monitoring Technology
2.1.2. Implantable Sensing Technology
- Glucose Monitoring: Implantable glucose sensors can be used to monitor blood sugar levels in people with diabetes [110]. These devices can continuously measure glucose levels and send data to a handheld device or smartphone, allowing patients to adjust their insulin dosages as needed.
- Neurological Monitoring: Implantable sensors can be used to monitor the brain activity in people with epilepsy, helping doctors to diagnose and treat the condition [112]. They can also be used to monitor intracranial pressure in people with traumatic brain injuries.
2.1.3. Ingestible Sensing Technology
- pH sensors are used to measure the acidity or alkalinity of the digestive system. These sensors can be used to diagnose conditions like acid reflux, gastroesophageal reflux disease (GERD), and Helicobacter pylori infection.
- Temperature sensors are used to measure the temperature of the digestive system. These sensors can be used to monitor body temperature and detect fever, as well as to diagnose conditions like Barrett’s esophagus and inflammatory bowel disease.
- Pressure sensors are used to measure the pressure within the digestive system. These sensors can be used to diagnose conditions like gastroparesis, achalasia, and other motility disorders.
- Electrolyte sensors are used to measure the levels of various electrolytes within the body, including sodium, potassium, and chloride. These sensors can be used to monitor electrolyte imbalances and diagnose conditions like dehydration and electrolyte disorders.
- Glucose sensors are used to measure blood sugar levels within the body. These sensors are commonly used to monitor glucose levels in people with diabetes.
- Drug sensors are used to monitor the absorption and distribution of medications within the body. These sensors can be used to optimize drug formulations and dosages for better treatment outcomes.
- Magnetic sensors are used to detect the presence of magnetic particles within the digestive system. These sensors can be used to diagnose conditions like gastrointestinal bleeding.
2.1.4. Smartphones
2.1.5. Others
2.2. Nonportable Sensing Technologies
- Stationary medical imaging technologies: Imaging technologies are noninvasive methods to visualize internal organs and diagnose various diseases [135]. Examples include X-ray, computed tomography (CT), magnetic resonance imaging (MRI), and positron emission tomography (PET). Owing to the extensive literature available on medical imaging methods and their applications in detecting and diagnosing various diseases and abnormalities, we have not provided detailed features of each method. Instead, we have referenced key review articles, such as Hosny et al., which presented a comprehensive overview of imaging technologies that have been enhanced with artificial intelligence techniques to diagnose various diseases [136]. Guluma et al. also reviewed DL methods in the detection of cancers using medical imaging data [137]. Additionally, Rana et al. discussed the use of ML and DL as medical imaging analysis tools for disease detection and diagnosis [138]. These articles provide valuable insights into the types of medical imaging data and applications of advanced computational techniques in medical imaging, and demonstrate their potential in improving disease diagnosis and patient outcomes.
- Environmental sensing technologies: They are used to detect and monitor environmental factors that can impact health conditions. Examples include air quality sensors, temperature sensors, and humidity sensors [139]. These sensors are used in smart homes. By combining these sensors with other DH technologies, they can play significant roles in improving the quality of care, reducing healthcare costs, and enhancing the independence and well-being of individuals [140].
- Monitoring and diagnostic technologies: Monitoring and diagnostic technologies based on biosensors are used to monitor and diagnose health conditions [141]. These devices are used to measure various biomarkers, such as glucose, cholesterol, and other vital signs, such as ECG, EEG, electro-oculography (EOG), and electroretinography (ERG).
- Robotic surgery systems: They are advanced medical devices that utilize robotic arms and computer-controlled instruments to assist surgeons in performing minimally invasive surgeries [141,142,143]. Examples of common robotic surgery systems include: (1) the da Vinci Surgical System [141], which is comprised of a console for the surgeon, and several robotic arms that hold surgical instruments and a camera; (2) MAKOplasty [142], utilized for orthopedic surgeries, such as knee and hip replacements; (3) the CyberKnife [143], employed for radiation therapy to treat cancer; (4) the ROSA Surgical System, utilized for neurosurgery procedures.
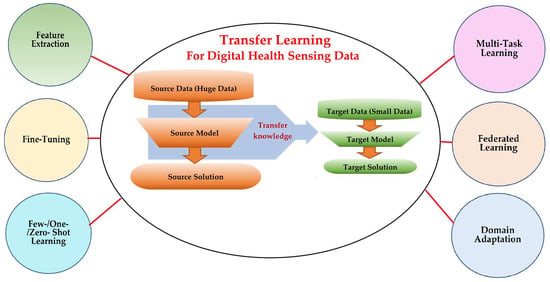
3. Transfer Learning: Strategies and Categories
3.1. Why the Transfer Learning Technique
- Appropriate modeling algorithms: there are many different types of ML algorithms, and choosing the right modeling algorithm for a particular task requires careful consideration of the data, the problem, and the desired outcome.
- Hyperparameter tuning: each ML method has hyperparameters that must be set before training, such as the learning rate, regularization strength, number of layers, etc. Determining the optimal values for these hyperparameters can be time-consuming, as it often requires many attempts to attain the best configuration.
- Data quality and privacy: preparing data to train ML models often requires extensive preprocessing of the raw data to enhance its quality and size. This involves techniques like normalization, scaling, transformation, feature selection, data augmentation, and data denoising, which demand careful considerations of the underlying data and the specific problem.
- Significant hardware resources: DL algorithms particularly require significant computational resources, including powerful GPUs, high-speed storage, and large amounts of memory, to perform complex computations due to the deep architectures that consist of various types of numerous kernels and layers. Several challenges are associated with these requirements, such as cost, availability, scalability, energy consumption, maintenance, and upgrade requirements.
3.2. Categories and Techniques of Transfer Learning
3.3. What to Transfer?
- Instance transfer: The ideal solution in TL is to effectively reuse knowledge from one domain to enhance the performance in another domain. However, the direct reuse of data from the source domain in the target domain is typically not feasible. Instead, the focus is on specific data instances from the source domain that can be combined with target data to enhance the results. This process is known as inductive transfer. This approach assumes that particular data portions from the source domain can be repurposed through techniques like instance reweighting and importance sampling.
- Feature-representation transfer: The goal of this approach is to decrease the differences between domains and improve the accuracy by finding valuable feature representations that can be shared from the source to the target domains. The choice between supervised and unsupervised methods for feature-based transfers depends on whether labeled data are accessible or not.
- Parameter transfer: This approach operates under the assumption that models for related tasks have certain shared parameters or a common distribution of hyperparameters. Multitask learning, where both the source and target tasks are learned simultaneously, is used in parameter-based TL.
- Relational-knowledge transfer: In contrast to the above three methods, relational-knowledge transfer aims to address non-independent and identically distributed data (non-IID), where each subsample exhibits significant variation and does not accurately represent the overall dataset distribution.
4. Applications of Transfer Learning on Digital Health Sensing Technologies
4.1. Methods, Strategies, and Applications of Transfer Learning in Digital Healthcare
4.1.1. Feature Extraction
4.1.2. Fine-Tuning
- Partial Fine-Tuning (unfreezing some layers)
- 2.
- Fully Fine-Tuning (unfreezing entire extracted layers)
- 3.
- Progressive Fine-Tuning (partially unfreezing the layers and training them on a multistage)
- 4.
- Adaptive Fine-Tuning (differentiating the learning rates for layer groups)
4.1.3. Domain Adaptation
4.1.4. Multitask Learning
4.1.5. Zero-Shot, One-Shot, and Few-Shot Learning
4.1.6. Federated Learning
4.2. Advantages and Disadvantages of Transfer Learning
- Improved performance: TL can help improve the performance of ML models, especially in cases where the training data are limited.
- Reduced training time: TL can reduce the amount of time and resources required to train an ML model, as the pretrained model can provide a starting point for learning.
- Reduced need for large datasets: TL can help mitigate the need for large datasets, as the pretrained model can provide a starting point for learning on smaller datasets.
- Increased generalization: TL can help improve the generalization of ML models, as the pretrained model has already learned the general features that can be applied to new datasets.
- Maintain data privacy: Multiple centers can collaboratively develop a global model without the need to share data to protect data sharing privacy.
- Domain-specific knowledge [239,240]: TL requires domain-specific knowledge to be effective. For example, if the Ds is image data, while the Dt is sound data, it is obvious that their features and distributions are dissimilar. Without finding a way to connect these two different domains, TL cannot be feasible.
- Limited flexibility: If the source task and target task are different and not related, it may not be easy to adapt the source task to a new task.
- Risk of negative transfer: TL can lead to a negative transfer for various reasons: distinct domains, conflicting assumptions, incompatible features, unbalanced transfer (if the source domain dominates the target domain, the model might be overfit to the source domain’s characteristics, leading to poor generalization on the target task), and model complexity. Additionally, transferring knowledge from noisy and limited source data cannot lead to positive outcomes in the target data.
- Limited interpretability: TL can make it more challenging to interpret the features learned by the model, as they may be influenced by the source model and may not necessarily be relevant to the target domain.
5. Conclusions and Future Work
- Adaptive Learning for real-time DH sensing Data:
- 2.
- Enabling TL on Edge Devices (EDs) for timely healthcare applications:
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Technology | Worn/Attached Location | Features and Applications |
|---|---|---|
| Smart watches and fitness trackers (Wearable) | Wrist, upper arm, waist, and ankle | Track physical activity, heart rate, sleep patterns, and other health metrics. |
| Smart lenses (Wearable) | Head/eye | Embedded with sensors to monitor glucose levels and other parameters in the tears, and send the data to a connected device. |
| Mouthguards (Wearable) | Head/mouth | Monitor various health metrics, such as the heart rate, breathing rate, and oxygen saturation, by measuring changes in the saliva and oral fluids. |
| Continuous glucose monitoring and insulin pumps (Wearable/attached to the skin using adhesive patches) | Abdomen (belly), upper buttocks, upper arm, and thigh | Help people with diabetes manage their blood sugar levels by continuously monitoring glucose levels and automatically delivering insulin as needed. |
| Headbands and hats (Wearable) | Head/around the forehead, and over the ears | Measure the brain activity, heart rate, and other vital signs. |
| Chest straps and attached bands (Wearable) | Around the chest or the wrist | Measure the heart rate variability, respiratory rate, and other health metrics by sensing changes in the skin conductivity or other physiological parameters used in the field of fitness and wellness. |
| ECG patches (Attached to the skin using a medical-grade adhesive) | Chest, upper back, or upper arm | It can monitor the heart rate, rhythm, and other cardiac metrics, and are often used to diagnose arrhythmias and other heart conditions. |
| Blood pressure cuffs (Wearable) | Wrist or upper arm | Measure blood pressure and can help diagnose and manage hypertension and other cardiovascular conditions. Some of these devices contain memory to store measurements and send information wirelessly to healthcare providers. |
| Smart clothing shirts, pants, and sports bras (Wearable) | Human body | Embedded with sensors to monitor vital signs, physical activity, and other health parameters. |
| Wearable cameras | Capture images and video of a patient’s environment, which can be used for telemedicine and remote monitoring purposes. | |
| Smart jewelry rings, bracelets, and necklaces (Wearable) | Different body locations | Equipped with sensors to track various health metrics. |
| Smart shoes (Wearable) | Foot | Detect gait patterns, track steps, and monitor posture. |
| Skin patches (Attachable) | Skin | Attached to the skin to monitor various physiological parameters, such as the heart rate, blood glucose levels, temperature, and hydration. |
| Smart helmets (Wearable) | Head | Enhance safety and provide connectivity. Equipped with sensors to monitor head impact forces and detect signs of concussion in athletes. |
| Sensor (Type of Data) | Features | Applications |
|---|---|---|
| Electrocardiogram (ECG) (Time series) | Measures electrical signals of the heart over time | Detecting arrhythmias and predicting heart disease [144,145] |
| Blood glucose monitoring (Time series) | Measures glucose levels over time | Predicting blood glucose levels [146,147] |
| Pulse oximeter (Time series) | Measures oxygen saturation and heart rate over time | Monitoring patients with respiratory (chronic obstructive pulmonary disease [148], COVID-19 [149], cardiac conditions [150]) |
| Electroencephalogram (EEG) (Time series) | Measures electrical activity in the brain over time | Predicting epilepsy [151], seizure risk [152], and diagnosing neurological disorders [153,154] |
| Accelerometer (Time series) | Measures movement over time | Monitoring physical activity [35,37,49] and predicting falls [155,156] |
| Blood pressure monitor (MEMS) (Time series) | Measures blood pressure over time | Cardiovascular monitoring [157] |
| X-ray (Image) | Images of internal structures, such as bones or organs | Diagnosing internal injuries or diseases (i.e., coronavirus [158,159], heart diseases [160]) |
| MRI (Image) | Images of internal structures, such as the brain or joints | Diagnosing internal injuries or diseases (i.e., cardiovascular diseases [161], cancers [162,163], knee injuries [164]) |
| CT scan (Image) | Images of internal structures, such as the brain or abdomen | Diagnosing internal injuries and diseases (i.e., cancers [165,166], cerebral aneurysm [167], lung diseases [168], and brain injuries [169]) |
| Ultrasound (Image) | Images of internal structures, such as the fetus or organs | Diagnosing internal injuries or diseases (i.e., carpal tunnel [170], liver diseases [171,172], and kidney injuries [173]) |
| Spirometer (Time series) | Measures lung function, including volume and flow rates | Predicting respiratory disease progression and monitoring the response to treatment [174,175] |
| Photoplethysmography (PPG) (Time series) | Measures various physiological parameters (heart rate, blood oxygen saturation, blood pressure, glucose levels, and emotional state) | Predicting glucose levels in patients with diabetes [176,177] and monitoring the emotional state or stress levels [178,179] |
| Electro-oculogram (EOG) (Time series) | Measures electrical signals from eye muscles and movements | Monitoring sleep patterns [180,181] and predicting eye disorder [182] |
| Infrared thermometer (Time series) | Measures body temperature from a distance | Monitoring patients with fever or hypothermia [183,184] |
| Optical coherence tomography (OCT) (Image) | Images of internal structures, such as the retina or cornea | Diagnosing eye diseases [185,186] |
| Capsule endoscope (Video) | Images and videos of the digestive tract | Diagnosing gastrointestinal disorders [187,188] |
| Acoustic (Video) | Measures acoustic features of the voice, e.g., the pitch, volume, and tone | Predicting Parkinson’s disease [189,190], diagnosing voice disorders [191], and detecting cardiac diseases [192] |
| Electrodermal activity sensor (EDA) (Time series) | Measures the electrical activity of sweat glands | Predicting emotional or psychological states [193,194] and monitoring stress levels [195] |
| Magnetometer (Time series) | Measures magnetic fields in the body | Monitoring cardiac function [196] and detecting locomotion and daily activities [197] |
| Photoacoustic imaging (Image) | Combines optical and ultrasound imaging for high-resolution images | Diagnosing cancer [198,199] and brain diseases [200] |
| Smart clothing (Time series) | Monitors vital signs and activity levels through sensors woven into clothing | Monitoring sleep [85], human motion [88], and detecting cardiovascular diseases [86]. |
| Pulse oximeter (Time series) | Measures oxygen saturation in the blood through a sensor on a finger or earlobe | Monitoring patients with respiratory [201] or cardiac conditions [202] |
| Multi-sensors (Multimodal signals data) (Time series) | Selects a few data features for better performance and higher accuracy | Multitask emotion recognition (valence, arousal, dominance, and liking) after watching videos [45] |
| Multi-sensors (Multimodal imaging data) (Image) | Provides information about tissues and internal organs, and functional information about metapolicy activities | Early detections of COVID-19 to assign appropriate treatment plans [203] |
Appendix B
| Task, Goal, and ML/DL Software to Develop the Model | Data Characteristics | Development Procedure | Achievements |
|---|---|---|---|
| Task: Automatically classify patients’ X-ray images into one of three categories: COVID-19, normal, and pneumonia. Goal: Overcome the difficulty in selecting the optimal engineering features to develop a reliable prediction model. Reduce the high dimensionality of the raw data, and improve its meaning. Software: Not specified. | Source: COVID-19 radiography database (open access provided by Kaggle). This database consists of 4 datasets:
|
|
|
| Task, Goal, and ML/DL Software to Develop the Model | Data Characteristics | Development Procedure | Achievements |
|---|---|---|---|
| Task: Classify human activities based on smartphone sensor data. Goal: Use fine-tuning to speed up training processes, overcome overfitting, and achieve a high classification accuracy in a new target task. Software: Not specified. | Source: Two state-of-the-art datasets were used: the “Khulna University Human Activity Recognition (KU-HAR)” and “the University of California Irvine Human Activities and Postural Transitions (UCI-HAPT)” Data usage:
KU-HAR dataset (2021): Contains 20,750 samples of 18 different activities (stand, sit, talk–sit, talk–stand, stand–sit, lay, lay–stand, pick, jump, push-up, sit-up, walk, walk backwards, walk–circle, run, stair–up, stair–down, and table tennis). Each sample lasted 3 s. The data were collected using a smartphone’s accelerometer and gyroscope sensors, worn at the waist. The data were gathered from 90 people aged 18 to 34. The data were not cleaned or filtered in any way so to consider a realistic dataset of real-world conditions. The dataset is unbalanced, and no data samples overlap with each other. UCI-HAPT (2014): Contains 10,929 samples collected from 30 volunteers (aged 19–48) using a waist-mounted smartphone triaxial accelerometer and gyroscope at the sampling rate of 50 Hz. It contains 12 activities (walking, walking upstairs, walking downstairs, sitting, standing, laying, stand-to-sit, sit-to-stand, sit-to-lie, lie-to-sit, stand-to-lie, and lie-to-stand). |
|
|
| Task, Goal, and ML/DL Software to Develop the Model | Data Characteristics | Development Procedure | Achievements |
|---|---|---|---|
| Task: Automatically classify patients’ MRI scans into one of three brain tumors: meningioma, glioma, and pituitary tumors, and segment the tumor regions from the MRI scans. Goal: Reduce development processes and improve the performance by jointly training two distinct but related tasks. Software: Not specified. | Source: Figshare MRI dataset. This dataset consists of: 3064 2D T1-weighted contrast-enhanced modalities (coronal, axial, and sagittal) collected from 233 patients. The classification and segmentation labels are included. Datadistribution:
|
|
|
References
- Gentili, A.; Failla, G.; Melnyk, A.; Puleo, V.; Tanna, G.L.D.; Ricciardi, W.; Cascini, F. The Cost-Effectiveness of Digital Health Interventions: A Systematic Review of The Literature. Front. Public Health 2022, 10, 787135. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, A.; Li, J.; Hardie, R.A.; Wabe, N.; Horvath, A.R.; Post, J.J.; Eigenstetter, A.; Lindeman, R.; Lam, Q.; Badrick, T.; et al. Diagnostic Informatics-The Role of Digital Health in Diagnostic Stewardship and the Achievement of Excellence, Safety, and Value. Front. Digit. Health 2021, 3, 659652. [Google Scholar] [CrossRef]
- Jagadeeswari, V.; Subramaniyaswamy, V.; Logesh, R.; Vijayakumar, V. A Study on Medical Internet of Things and Big Data in Personalized Healthcare System. Health Inf. Sci. Syst. 2018, 6, 14. [Google Scholar] [CrossRef] [PubMed]
- Mbunge, E.; Muchemwa, B.; Jiyane, S.; Batani, J. Sensors and Healthcare 5.0: Transformative Shift in Virtual Care Through Emerging Digital Health Technologies. Glob. Health J. 2021, 5, 169–177. [Google Scholar] [CrossRef]
- Butcher, C.J.; Hussain, W. Digital Healthcare: The Future. Future Healthc. J. 2022, 9, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Thompson, C.A.; Peterson, S.K.; Mandrola, J.; Beg, M.S. The Future of Wearable Technologies and Remote Monitoring in Health Care. Am. Soc. Clin. Oncol. Educ. Book 2019, 39, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Vesnic-Alujevic, L.; Breitegger, M.; Guimarães Pereira, Â. ‘Do-It-Yourself’ Healthcare? Quality of Health and Healthcare Through Wearable Sensors. Sci. Eng. Ethics 2018, 24, 887–904. [Google Scholar] [CrossRef]
- Anikwe, C.V.; Nweke, H.F.; Ikegwu, A.C.; Egwuonwu, C.A.; Onu, F.U.; Alo, U.R.; Teh, Y.W. Mobile and Wearable Sensors for Data-Driven Health Monitoring System: State-of-The-Art and Future Prospect. Expert Syst. Appl. 2022, 202, 117362. [Google Scholar] [CrossRef]
- Verma, P.S.; Sood, S.K. Cloud-Centric IoT based Disease Diagnosis Healthcare Framework. J. Parallel Distrib. Comput. 2018, 116, 27–38. [Google Scholar] [CrossRef]
- Rutledge, C.M.; Kott, K.; Schweickert, P.A.; Poston, R.; Fowler, C.; Haney, T.S. Telehealth and eHealth in Nurse Practitioner Training: Current Perspectives. Adv. Med. Educ. Pract. 2017, 8, 399–409. [Google Scholar] [CrossRef]
- Guk, K.; Han, G.; Lim, J.; Jeong, K.; Kang, T.; Lim, E.K.; Jung, J. Evolution of Wearable Devices with Real-Time Disease Monitoring for Personalized Healthcare. Nanomaterials 2019, 9, 813. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Imani, S.; de Araujo, W.R.; Warchall, J.; Valdés-Ramírez, G.; Paixão, T.R.L.C.; Mercier, P.P.; Wang, J. Wearable Salivary Uric Acid Mouthguard Biosensor with Integrated Wireless Electronics. Biosens. Bioelectron. 2015, 74, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Campbell, A.S.; Wang, J. Wearable Non-Invasive Epidermal Glucose Sensors: A Review. Talanta 2018, 177, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Viswanath, B.; Choi, C.S.; Lee, K.; Kim, S. Recent Trends in the Development of Diagnostic Tools for Diabetes Mellitus using Patient Saliva. Trends Anal. Chem. 2017, 89, 60–67. [Google Scholar] [CrossRef]
- Arakawa, T.; Tomoto, K.; Nitta, H.; Toma, K.; Takeuchi, S.; Sekita, T.; Minakuchi, S.; Mitsubayashi, K. A Wearable Cellulose Acetate-Coated Mouthguard Biosensor for In Vivo Salivary Glucose Measurement. Anal. Chem. 2020, 92, 12201–12207. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.L.; Ding, X.R.; Poon, C.C.Y.; Lo, B.P.L.; Zhang, H.; Zhou, X.L.; Yang, G.Z.; Zhao, N.; Zhang, Y.T. Unobtrusive Sensing and Wearable Devices for Health Informatics. IEEE Trans. Biomed. Eng. 2014, 61, 1538–1554. [Google Scholar] [CrossRef] [PubMed]
- Sempionatto, J.R.; Brazaca, L.C.; García-Carmona, L.; Bolat, G.; Campbell, A.S.; Martin, A.; Tang, G.; Shah, R.; Mishra, R.K.; Kim, J.; et al. Eyeglasses-Based Tear Biosensing System: Non-Invasive Detection of Alcohol, Vitamins and Glucose. Biosens. Bioelectron. 2019, 137, 161–170. [Google Scholar] [CrossRef]
- Constant, N.; Douglas-Prawl, O.; Johnson, S.; Mankodiya, K. Pulse-Glasses: An Unobtrusive, Wearable HR Monitor with Internet-of-Things Functionality. In Proceedings of the 2015 IEEE 12th International Conference on Wearable and Implantable Body Sensor Networks (BSN), Cambridge, MA, USA, 9–12 June 2015; pp. 1–5. [Google Scholar] [CrossRef]
- Liu, X.; Lillehoj, P.B. Embroidered Electrochemical Sensors for Biomolecular Detection. Lab A Chip 2016, 16, 2093–2098. [Google Scholar] [CrossRef]
- Jung, P.G.; Lim, G.; Kong, K. A Mobile Motion Capture System based on Inertial Sensors and Smart Shoes. J. Dyn. Syst. Meas. Control 2014, 136, 011002. [Google Scholar] [CrossRef]
- Kim, J.H.; Roberge, R.; Powell, J.B.; Shafer, A.B.; Williams, W.J. Measurement Accuracy of Heart Rate and Respiratory Rate During Graded Exercise and Sustained Exercise in the Heat Using the Zephyr BioHarnessTM. Int. J. Sports Med. 2013, 34, 497–501. [Google Scholar] [CrossRef]
- Mihai, D.A.; Stefan, D.S.; Stegaru, D.; Bernea, G.E.; Vacaroiu, I.A.; Papacocea, T.; Lupușoru, M.O.D.; Nica, A.E.; Stiru, O.; Dragos, D.; et al. Continuous Glucose Monitoring Devices: A Brief Presentation (Review). Exp. Ther. Med. 2022, 23, 174. [Google Scholar] [CrossRef] [PubMed]
- Gamessa, T.W.; Suman, D.; Tadesse, Z.K. Blood Glucose Monitoring Techniques: Recent Advances, Challenges and Future Perspectives. Int. J. Adv. Technol. Eng. Explor. 2018, 5, 335–344. [Google Scholar] [CrossRef]
- Javaid, M.; Haleem, A.; Singh, R.P.; Suman, R.; Rab, S. Significance of Machine Learning in Healthcare: Features, Pillars and Applications. Int. J. Intell. Netw. 2022, 3, 58–73. [Google Scholar] [CrossRef]
- Qayyum, A.; Qadir, J.; Bilal, M.; Al-Fuqaha, A. Secure and Robust Machine Learning for Healthcare: A Survey. IEEE Rev. Biomed. Eng. 2021, 14, 156–180. [Google Scholar] [CrossRef]
- Islam, M.S.; Hasan, M.M.; Wang, X.; Germack, H.D.; Noor-E-Alam, M. A Systematic Review on Healthcare Analytics: Application and Theoretical Perspective of Data Mining. Healthcare 2018, 6, 54. [Google Scholar] [CrossRef] [PubMed]
- Bohr, A.; Memarzadeh, K. The Rise of Artificial Intelligence in Healthcare Applications. Artif. Intell. Healthc. 2020, 25–60. [Google Scholar] [CrossRef]
- Love-Koh, J.; Peel, A.; Rejon-Parrilla, J.C.; Ennis, K.; Lovett, R.; Manca, A.; Taylor, M. The Future of Precision Medicine: Potential Impacts for Health Technology Assessment. Pharmacoeconomics 2018, 36, 1439–1451. [Google Scholar] [CrossRef] [PubMed]
- Berger, J.S.; Haskell, L.; Ting, W.; Lurie, F.; Chang, S.C.; Mueller, L.A.; Elder, K.; Rich, K.; Crivera, C.; Schein, J.R.; et al. Evaluation of Machine Learning Methodology for the Prediction of Healthcare Resource Utilization and Healthcare Costs in Patients With Critical Limb Ischemia-Is Preventive and Personalized Approach on the Horizon? EPMA J. 2020, 11, 53–64. [Google Scholar] [CrossRef]
- Kelly, C.J.; Karthikesalingam, A.; Suleyman, M.; Corrado, G.; King, D. Key Challenges for Delivering Clinical Impact with Artificial Intelligence. BMC Med. 2019, 17, 195. [Google Scholar] [CrossRef]
- Ellis, R.J.; Sander, R.; Limon, A. Twelve Key Challenges in Medical Machine Learning and Solutions. Intell.-Based Med. 2022, 6, 100068. [Google Scholar] [CrossRef]
- Hosna, A.; Merry, E.; Gyalmo, J.; Alom, Z.; Aung, Z.; Azim, M.A. Transfer Learning: A Friendly Introduction. J. Big Data 2022, 9, 102. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.J.; Yang, Q. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 2010, 22, 1345–1359. [Google Scholar] [CrossRef]
- Zhuang, F.; Qi, Z.; Duan, K.; Xi, D.; Zhu, Y.; Zhu, H.; Xiong, H.; He, Q. A Comprehensive Survey on Transfer Learning. Proc. IEEE 2021, 109, 43–76. [Google Scholar] [CrossRef]
- Pavliuk, O.; Mishchuk, M.; Strauss, C. Transfer Learning Approach for Human Activity Recognition Based on Continuous Wavelet Transform. Algorithms 2023, 16, 77. [Google Scholar] [CrossRef]
- An, S.; Bhat, G.; Gümüşsoy, S.; Ogras, Ü.Y. Transfer Learning for Human Activity Recognition Using Representational Analysis of Neural Networks. ACM Trans. Comput. Healthc. 2023, 4, 1–21. [Google Scholar] [CrossRef]
- Waters, S.H.; Clifford, G.D. Comparison of Deep Transfer Learning Algorithms and Transferability Measures for Wearable Sleep Staging. Biomed. Eng. Online 2022, 21, 66. [Google Scholar] [CrossRef] [PubMed]
- Abou Jaoude, M.; Sun, H.; Pellerin, K.R.; Pavlova, M.; Sarkis, R.A.; Cash, S.S.; Westover, M.B.; Lam, A.D. Expert-Level Automated Sleep Staging of Long-Term Scalp Electroencephalography Recordings using Deep Learning. Sleep 2020, 43, zsaa112. [Google Scholar] [CrossRef]
- Li, Q.; Li, Q.; Cakmak, A.S.; Da Poian, G.; Bliwise, D.L.; Vaccarino, V.; Shah, A.J.; Clifford, G.D. Transfer learning from ECG to PPG for improved sleep staging from wrist-worn wearables. Physiol. Meas. 2021, 42, 044004. [Google Scholar] [CrossRef]
- Radha, M.; Fonseca, P.; Moreau, A.; Ross, M.; Cerny, A.; Anderer, P.; Long, X.; Aarts, R.M. A Deep Transfer Learning Approach for Wearable Sleep Stage Classification with Photoplethysmography. npj Digit. Med. 2021, 4, 135. [Google Scholar] [CrossRef]
- Narin, A. Accurate detection of COVID-19 Using Deep Features based on X-Ray Images and Feature Selection Methods. Comput. Biol. Med. 2021, 137, 104771. [Google Scholar] [CrossRef]
- Salem, M.A.; Taheri, S.; Yuan, J. ECG Arrhythmia Classification Using Transfer Learning from 2- Dimensional Deep CNN Features. In Proceedings of the 2018 IEEE Biomedical Circuits and Systems Conference (BioCAS), Cleveland, OH, USA, 17–19 October 2018; pp. 1–4. [Google Scholar]
- Vo, D.M.; Nguyen, N.Q.; Lee, S.W. Classification of Breast Cancer Histology Images using Incremental Boosting Convolution Networks. Inf. Sci. 2019, 482, 123–138. [Google Scholar] [CrossRef]
- Thuy, M.B.H.; Hoang, V.T. Fusing of Deep Learning, Transfer Learning and GAN for Breast Cancer Histopathological Image Classification. In Advanced Computational Methods for Knowledge Engineering: Proceedings of the 6th International Conference on Computer Science, Applied Mathematics and Applications, ICCSAMA 2019; Springer International Publishing: Berlin/Heidelberg, Germany, 2020; Volume 6, pp. 255–266. [Google Scholar] [CrossRef]
- He, X.; Huang, J.; Zeng, Z. Logistic Regression Based Multi-task, Multi-kernel Learning for Emotion Recognition. In Proceedings of the 2021 6th IEEE International Conference on Advanced Robotics and Mechatronics (ICARM), Chongqing, China, 3–5 July 2021; pp. 572–577. [Google Scholar] [CrossRef]
- Caruana, R. Multitask Learning. In Learning to Learn; Thrun, S., Pratt, L., Eds.; Springer: Boston, MA, USA, 1998. [Google Scholar] [CrossRef]
- Wang, J.; Chen, Y.; Hu, L.; Peng, X.; Philip, S.Y. Stratified Transfer Learning for Cross-Domain Activity Recognition. In Proceedings of the 2018 IEEE international conference on pervasive computing and communications (PerCom), Athens, Greece, 19–23 March 2018; pp. 1–10. [Google Scholar] [CrossRef]
- Chakma, A.; Faridee, A.Z.M.; Khan, M.A.A.H.; Roy, N. Activity Recognition in Wearables Using Adversarial Multi-Source Domain Adaptation. Smart Health 2021, 19, 100174. [Google Scholar] [CrossRef]
- Presotto, R.; Civitarese, G.; Bettini, C. Semi-Supervised and Personalized Federated Activity Recognition based on Active Learning and Label Propagation. Pers. Ubiquitous Comput. 2022, 26, 1281–1298. [Google Scholar] [CrossRef]
- Chen, Y.; Qin, X.; Wang, J.; Yu, C.; Gao, W. FedHealth: A Federated Transfer Learning Framework for Wearable Healthcare. IEEE Intell. Syst. 2020, 35, 83–93. [Google Scholar] [CrossRef]
- Zhang, A.S.; Li, N.F. When Accuracy Meets Privacy: Two-Stage Federated Transfer Learning Framework in Classification of Medical Images on Limited Data: A COVID-19 Case Study. arXiv 2022, arXiv:2203.12803. [Google Scholar] [CrossRef]
- Mishra, N.; Rohaninejad, M.; Chen, X.; Abbeel, P. A Simple Neural Attentive Meta-Learner. arXiv 2018, arXiv:1707.03141. [Google Scholar]
- Liu, T.; Yang, Y.; Fan, W.; Wu, C. Reprint of: Few-Shot Learning for Cardiac Arrhythmia Detection based on Electrocardiogram Data from Wearable Devices. Digit. Signal Process. 2022, 125, 103574. [Google Scholar] [CrossRef]
- Zhang, P.; Li, J.; Wang, Y.; Pan, J. Domain Adaptation for Medical Image Segmentation: A Meta-Learning Method. J. Imaging 2021, 7, 31. [Google Scholar] [CrossRef]
- Xu, S.; Kim, J.; Walter, J.R.; Ghaffari, R.; Rogers, J.A. Translational Gaps and Opportunities for Medical Wearables in Digital Health. Sci. Transl. Med. 2022, 14, eabn6036. [Google Scholar] [CrossRef]
- Ullah, F.; Haq, H.; Khan, J.; Safeer, A.A.; Asif, U.; Lee, S. Wearable IoTs and Geo-Fencing Based Framework for COVID-19 Remote Patient Health Monitoring and Quarantine Management to Control the Pandemic. Electronics 2021, 10, 2035. [Google Scholar] [CrossRef]
- Sardar, A.W.; Ullah, F.; Bacha, J.; Khan, J.; Ali, F.; Lee, S. Mobile Sensors based Platform of Human Physical Activities Recognition for COVID-19 spread minimization. Comput. Biol. Med. 2022, 146, 105662. [Google Scholar] [CrossRef] [PubMed]
- Papini, G.B.; Fonseca, P.; Van Gilst, M.M.; Bergmans, J.W.M.; Vullings, R.; Overeem, S. Wearable Monitoring of Sleep-Disordered Breathing: Estimation of the Apnea–Hypopnea index using Wrist-Worn Reflective Photoplethysmography. Sci. Rep. 2020, 10, 13512. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ellul, J.; Azzopardi, G. Elderly Fall Detection Systems: A Literature Survey. Front. Robot. AI 2020, 7, 71. [Google Scholar] [CrossRef] [PubMed]
- Thilo, F.J.S.; Hahn, S.; Halfens, R.J.G.; Schols, J.M.G.A. Usability of a Wearable Fall Detection Prototype from the Perspective of Older People–A Real Field Testing Approach. J. Clin. Nurs. 2018, 28, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Konstantinidis, D.; Iliakis, P.; Tatakis, F.; Thomopoulos, K.; Dimitriadis, K.; Tousoulis, D.; Tsioufis, K. Wearable Blood Pressure Measurement Devices and New Approaches in Hypertension Management: The Digital Era. J. Hum. Hypertens. 2022, 36, 945–951. [Google Scholar] [CrossRef]
- Tran, A.; Zhang, X.; Zhu, B. Mechanical Structural Design of a Piezoresistive Pressure Sensor for Low-Pressure Measurement: A Computational Analysis by Increases in the Sensor Sensitivity. Sensors 2018, 18, 2023. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekhar, A.; Kim, C.S.; Naji, M.; Natarajan, K.; Hahn, J.O.; Mukkamala, R. Smartphone-Based Blood Pressure Monitoring Via the Oscillometric Finger-Pressing Method. Sci. Transl. Med. 2018, 10, eaap8674. [Google Scholar] [CrossRef] [PubMed]
- Jafarzadeh, F.; Rahmani, F.; Azadmehr, F.; Falaki, M.; Nazari, M. Different Applications of Telemedicine—Assessing the Challenges, Barriers, And Opportunities- A Narrative Review. J. Fam. Med. Prim. Care 2022, 11, 879–886. [Google Scholar] [CrossRef]
- Bouabida, K.; Lebouché, B.; Pomey, M.-P. Telehealth and COVID-19 Pandemic: An Overview of the Telehealth Use, Advantages, Challenges, and Opportunities during COVID-19 Pandemic. Healthcare 2022, 10, 2293. [Google Scholar] [CrossRef]
- Doniec, R.J.; Piaseczna, N.J.; Szymczyk, K.A.; Jacennik, B.; Sieciński, S.; Mocny-Pachońska, K.; Duraj, K.; Cedro, T.; Tkacz, E.J.; Glinkowski, W.M. Experiences of the Telemedicine and eHealth Conferences in Poland—A Cross-National Overview of Progress in Telemedicine. Appl. Sci. 2023, 13, 587. [Google Scholar] [CrossRef]
- Serhani, M.A.; EL Kassabi, H.T.; Ismail, H.; Nujum Navaz, A. ECG Monitoring Systems: Review, Architecture, Processes, and Key Challenges. Sensors 2020, 20, 1796. [Google Scholar] [CrossRef] [PubMed]
- Turakhia, M.P.; Hoang, D.D.; Zimetbaum, P.; Miller, J.D.; Froelicher, V.F.; Kumar, U.N.; Xu, X.; Yang, F.; Heidenreich, P.A. Heidenreich, Diagnostic Utility of a Novel Leadless Arrhythmia Monitoring Device. Am. J. Cardiol. 2013, 112, 520–524. [Google Scholar] [CrossRef] [PubMed]
- Braunstein, E.D.; Reynbakh, O.; Krumerman, A.; Di Biase, L.; Ferrick, K.J. Inpatient Cardiac Monitoring Using a Patch-Based Mobile Cardiac Telemetry System During The COVID-19 pandemic. J. Cardiovasc. Electrophysiol. 2020, 31, 2803–2811. [Google Scholar] [CrossRef] [PubMed]
- Gomes, N.; Pato, M.; Lourenço, A.R.; Datia, N. A Survey on Wearable Sensors for Mental Health Monitoring. Sensors 2023, 23, 1330. [Google Scholar] [CrossRef]
- Perna, G.; Riva, A.; Defillo, A.; Sangiorgio, E.; Nobile, M.; Caldirola, D. Heart Rate Variability: Can It Serve as a Marker of Mental Health Resilience? J. Affect. Disord. 2020, 263, 754–761. [Google Scholar] [CrossRef]
- Dobson, R.; Li, L.L.; Garner, K.; Tane, T.; McCool, J.; Whittaker, R. The Use of Sensors to Detect Anxiety for In-the-Moment Intervention: Scoping Review. JMIR Ment. Health 2023, 10, e42611. [Google Scholar] [CrossRef]
- Crupi, R.; Faetti, T.; Paradiso, R. Preliminary Evaluation of Wearable Wellness System for Obstructive Sleep Apnea Detection. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; pp. 4142–4145. [Google Scholar] [CrossRef]
- Chung, A.H.; Gevirtz, R.N.; Gharbo, R.S.; Thiam, M.A.; Ginsberg, J.P. Pilot Study on Reducing Symptoms of Anxiety with a Heart Rate Variability Biofeedback Wearable and Remote Stress Management Coach. Appl. Psychophysiol. Biofeedback 2021, 46, 347–358. [Google Scholar] [CrossRef]
- Morin, C.M.; Bjorvatn, B.; Chung, F.; Holzinger, B.; Partinen, M.; Penzel, T.; Ivers, H.; Wing, Y.K.; Chan, N.Y.; Merikanto, I.; et al. Insomnia, Anxiety, and Depression during the COVID-19 Pandemic: An International Collaborative Study. Sleep Med. 2021, 87, 38–45. [Google Scholar] [CrossRef]
- Can, Y.S.; Arnrich, B.; Ersoy, C. Stress Detection in Daily Life Scenarios Using Smart Phones and Wearable Sensors: A Survey. J. Biomed. Inform. 2019, 92, 103139. [Google Scholar] [CrossRef]
- Bunn, J.A.; Navalta, J.W.; Fountaine, C.J.; Reece, J.D. Current State of Commercial Wearable Technology in Physical Activity Monitoring 2015–2017. Int. J. Exerc. Sci. 2018, 11, 503–515. [Google Scholar]
- McGinnis, R.S.; Mcginnis, E.W.; Petrillo, C.J.; Price, M. Mobile Biofeedback Therapy for the Treatment of Panic Attacks: A Pilot Feasibility Study. In Proceedings of the 2019 IEEE 16th International Conference on Wearable and Implantable Body Sensor Networks (BSN), Chicago, IL, USA, 19–22 May 2019; pp. 1–4. [Google Scholar] [CrossRef]
- Hilty, D.M.; Armstrong, C.M.; Luxton, D.D.; Gentry, M.T.; Krupinski, E.A. A Scoping Review of Sensors, Wearables, and Remote Monitoring for Behavioral Health: Uses, Outcomes, Clinical Competencies, and Research Directions. J. Technol. Behav. Sci. 2021, 6, 278–313. [Google Scholar] [CrossRef]
- Kwon, S.; Kim, H.; Yeo, W.-H. Recent Advances in Wearable Sensors and Portable Electronics for Sleep Monitoring. iScience 2021, 24, 102461. [Google Scholar] [CrossRef]
- Liao, L.D.; Wang, Y.; Tsao, Y.C.; Wang, I.J.; Jhang, D.F.; Chuang, C.C.; Chen, S.F. Design and Implementation of a Multifunction Wearable Device to Monitor Sleep Physiological Signals. Micromachines 2020, 11, 672. [Google Scholar] [CrossRef] [PubMed]
- Pham, N.T.; Dinh, T.A.; Raghebi, Z.; Kim, T.; Bui, N.; Nguyen, P.; Truong, H.; Banaei-Kashani, F.; Halbower, A.C.; Dinh, T.N.; et al. WAKE: A Behind-The-Ear Wearable System for Microsleep Detection. In Proceedings of the 18th International Conference on Mobile Systems, Applications, and Services (MobiSys ′20), Association for Computing Machinery, New York, NY, USA, 15–19 June 2020; pp. 404–418. [Google Scholar] [CrossRef]
- Meng, K.; Zhao, S.; Zhou, Y.; Wu, Y.; Zhang, S.; He, Q.; Wang, X.; Zhou, Z.; Fan, W.; Tan, X.; et al. A Wireless Textile-Based Sensor System for Self-Powered Personalized Health Care. Matter 2020, 2, 896–907. [Google Scholar] [CrossRef]
- Fang, Y.; Zou, Y.; Xu, J.; Chen, G.; Zhou, Y.; Deng, W.; Zhao, X.; Roustaei, M.; Hsiai, T.K.; Chen, J. Ambulatory Cardiovascular Monitoring Via a Machine-Learning-Assisted Textile Triboelectric Sensor. Adv. Mater. 2021, 33, 2104178. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Pang, Y.; Han, X.; Yang, Y.; Ling, J.; Jian, M.; Zhang, Y.; Yang, Y.; Ren, T.L. Graphene Textile Strain Sensor with Negative Resistance Variation for Human Motion Detection. ACS Nano 2018, 12, 9134–9141. [Google Scholar] [CrossRef] [PubMed]
- Di Tocco, J.; Lo Presti, D.; Rainer, A.; Schena, E.; Massaroni, C. Silicone-Textile Composite Resistive Strain Sensors for Human Motion-Related Parameters. Sensors 2022, 22, 3954. [Google Scholar] [CrossRef] [PubMed]
- Vaddiraju, S.; Burgess, D.J.; Tomazos, I.; Jain, F.C.; Papadimitrakopoulos, F. Technologies for Continuous Glucose Monitoring: Current Problems and Future Promises. J. Diabetes Sci. Technol. 2010, 4, 1540–1562. [Google Scholar] [CrossRef]
- Dungan, K.; Verma, N. Monitoring Technologies—Continuous Glucose Monitoring, Mobile Technology, Biomarkers of Glycemic Control. In Endotext; MDText.com, Inc.: South Dartmouth, MA, USA, 2000. Available online: https://www.ncbi.nlm.nih.gov/books/NBK279046/ (accessed on 8 July 2023).
- Ma, X.; Ahadian, S.; Liu, S.; Zhang, J.; Liu, S.; Cao, T.; Lin, W.; Wu, D.; de Barros, N.R.; Zare, M.R.; et al. Smart Contact Lenses for Biosensing Applications. Adv. Intell. Syst. 2021, 3, 2000263. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, S.; Cui, Q.; Ni, J.; Wang, X.; Cheng, X.; Alem, H.; Tebon, P.; Xu, C.; Guo, C.; et al. Microengineered Poly (HEMA) Hydrogels for wearable Contact Lens Biosensing. Lab Chip 2020, 20, 4205–4214. [Google Scholar] [CrossRef]
- Gao, W.; Emaminejad, S.; Nyein, H.Y.Y.; Challa, S.; Chen, K.; Peck, A.; Fahad, H.M.; Ota, H.; Shiraki, H.; Kiriya, D.; et al. Fully Integrated Wearable Sensor Arrays for Multiplexed in Situ Perspiration Analysis. Nature 2016, 529, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Farandos, N.M.; Yetisen, A.K.; Monteiro, M.J.; Lowe, C.R.; Yun, S.H. Contact Lens Sensors in Ocular Diagnostics. Adv. Healthc. Mater. 2015, 4, 792–810. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Choi, T.K.; Lee, Y.B.; Cho, H.R.; Ghaffari, R.; Wang, L.; Choi, H.J.; Chung, T.D.; Lu, N.; Hyeon, T.; et al. A Graphene-Based Electrochemical Device with Thermoresponsive Microneedles for Diabetes Monitoring and Therapy. Nat. Nanotechnol. 2016, 11, 566–572. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Song, C.; Hong, Y.S.; Kim, M.; Cho, H.R.; Kang, T.; Shin, K.; Choi, S.H.; Hyeon, T.; Kim, D.H. Wearable/Disposable Sweat-Based Glucose Monitoring Device with Multistage Transdermal Drug Delivery Module. Sci. Adv. 2017, 3, e1601314. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, G.; Jeon, C.; Han, H.H.; Kim, S.; Mok, J.W.; Joo, C.; Shin, S.; Sim, J.; Myung, D.; et al. Bimetallic Nanocatalysts Immobilized in Nanoporous Hydrogels for Long-Term Robust Continuous Glucose Monitoring of Smart Contact Lens. Adv. Mater. 2022, 34, 2110536. [Google Scholar] [CrossRef] [PubMed]
- Jalloul, N. Wearable Sensors for The Monitoring of Movement Disorders. Biomed. J. 2018, 41, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.Z.; Soylu, A. Human Activity Recognition Using Wearable Sensors, Discriminant Analysis, And Long Short-Term Memory-Based Neural Structured Learning. Sci. Rep. 2021, 11, 16455. [Google Scholar] [CrossRef]
- Csizmadia, G.; Liszkai-Peres, K.; Ferdinandy, B.; Miklósi, Á.; Konok, V. Human Activity Recognition of Children with Wearable Devices Using Lightgbm Machine Learning. Sci. Rep. 2022, 12, 5472. [Google Scholar] [CrossRef]
- Pacchierotti, C.; Salvietti, G.; Hussain, I.; Meli, L.; Prattichizzo, D. The hRing: A Wearable Haptic Device to Avoid Occlusions in Hand Tracking. In Proceedings of the 2016 IEEE Haptics Symposium (HAPTICS), Philadelphia, PA, USA, 8–11 April 2016; pp. 134–139. [Google Scholar] [CrossRef]
- Fan, C.; Gao, F. Enhanced Human Activity Recognition Using Wearable Sensors via a Hybrid Feature Selection Method. Sensors 2021, 21, 6434. [Google Scholar] [CrossRef]
- Li, Y.; Tang, H.; Liu, Y.; Qiao, Y.; Xia, H.; Zhou, J. Oral Wearable Sensors: Health Management Based on the Oral Cavity. Biosens. Bioelectron. X 2022, 10, 100135. [Google Scholar] [CrossRef]
- Li, X.; Luo, C.; Fu, Q.; Zhou, C.; Ruelas, M.; Wang, Y.; He, J.; Wang, Y.; Zhang, Y.S.; Zhou, J. A Transparent, Wearable Fluorescent Mouthguard for High-Sensitive Visualization and Accurate Localization of Hidden Dental Lesion Sites. Adv. Mater. 2020, 32, e2000060. [Google Scholar] [CrossRef]
- Quadir, N.A.; Albasha, L.; Taghadosi, M.; Qaddoumi, N.; Hatahet, B. Low-Power Implanted Sensor for Orthodontic Bond Failure Diagnosis and Detection. IEEE Sens. J. 2018, 18, 3003–3009. [Google Scholar] [CrossRef]
- Bodini, A.; Borghetti, M.; Paganelli, C.; Sardini, E.; Serpelloni, M. Low-Power Wireless System to Monitor Tongue Strength Against the Palate. IEEE Sens. J. 2021, 21, 5467–5475. [Google Scholar] [CrossRef]
- Gawande, P.; Deshmukh, A.; Dhangar, R.; Gare, K.; More, S. A Smart Footwear System for Healthcare and Fitness Application—A Review. J. Res. Eng. Appl. Sci. 2020, 5, 10–14. [Google Scholar] [CrossRef]
- Mehendale, N.; Gokalgandhi, D.; Shah, N.; Kamdar, L. A Review of Smart Technologies Embedded in Shoes. SSRN Electron. J. 2020, 44, 150. [Google Scholar] [CrossRef]
- Bae, C.W.; Toi, P.T.; Kim, B.Y.; Lee, W.I.; Lee, H.B.; Hanif, A.; Lee, E.H.; Lee, N.-E. Fully Stretchable Capillary Microfluidics-Integrated Nanoporous Gold Electrochemical Sensor for Wearable Continuous Glucose Monitoring. ACS Appl. Mater. Interfaces 2019, 11, 14567–14575. [Google Scholar] [CrossRef]
- Lin, S.; Wang, B.; Zhao, Y.; Shih, R.; Cheng, X.; Yu, W.; Hojaiji, H.; Lin, H.; Hoffman, C.; Ly, D.; et al. Natural Perspiration Sampling and in Situ Electrochemical Analysis with Hydrogel Micropatches for User-Identifiable and Wireless Chemo/Biosensing. ACS Sens. 2019, 5, 93–102. [Google Scholar] [CrossRef]
- Darwish, A.; Hassanien, A.E. Wearable and Implantable Wireless Sensor Network Solutions for Healthcare Monitoring. Sensors 2011, 11, 5561–5595. [Google Scholar] [CrossRef]
- Dinis, H.; Mendes, P.M. Recent Advances on Implantable Wireless Sensor Networks. Wirel. Sens. Netw. Insights Innov. 2017. [Google Scholar] [CrossRef]
- Klosterhoff, B.S.; Tsang, M.; She, D.; Ong, K.G.; Allen, M.G.; Willett, N.J.; Guldberg, R.E. Implantable Sensors for Regenerative Medicine. J. Biomech. Eng. 2017, 139, 021009. [Google Scholar] [CrossRef]
- McShane, M.J.; Zavareh, A.T.; Jeevarathinam, A.S. Implantable Sensors. Encycl. Sens. Biosens. 2023, 4, 115–132. [Google Scholar] [CrossRef]
- Bisignani, A.; De Bonis, S.; Mancuso, L.; Ceravolo, G.; Bisignani, G. Implantable Loop Recorder in Clinical Practice. J. Arrhythmia 2018, 35, 25–32. [Google Scholar] [CrossRef]
- Talebian, S.; Foroughi, J.; Wade, S.J.; Vine, K.L.; Dolatshahi-Pirouz, A.; Mehrali, M.; Conde, J.; Wallace, G.G. Biopolymers for Antitumor Implantable Drug Delivery Systems: Recent Advances and Future Outlook. Adv. Mater. 2018, 30, e1706665. [Google Scholar] [CrossRef]
- Pial, M.M.H.; Tomitaka, A.; Pala, N.; Roy, U. Implantable Devices for the Treatment of Breast Cancer. J. Nanotheranostics 2022, 3, 19–38. [Google Scholar] [CrossRef]
- Chong, K.P.; Woo, B.K. Emerging wearable technology applications in gastroenterology: A Review of the Literature. World J. Gastroenterol. 2021, 27, 1149–1160. [Google Scholar] [CrossRef]
- Beardslee, L.A.; Banis, G.E.; Chu, S.; Liu, S.; Chapin, A.A.; Stine, J.M.; Pasricha, P.J.; Ghodssi, R. Ingestible Sensors and Sensing Systems for Minimally Invasive Diagnosis and Monitoring: The Next Frontier in Minimally Invasive Screening. ACS Sens. 2020, 5, 891–910. [Google Scholar] [CrossRef]
- Dagdeviren, C.; Javid, F.; Joe, P.; von Erlach, T.; Bensel, T.; Wei, Z.; Saxton, S.; Cleveland, C.; Booth, L.; McDonnell, S.; et al. Flexible Piezoelectric Devices for Gastrointestinal Motility Sensing. Nat. Biomed. Eng. 2017, 1, 807–817. [Google Scholar] [CrossRef]
- Mimee, M.; Nadeau, P.; Hayward, A.; Carim, S.; Flanagan, S.; Jerger, L.; Collins, J.; McDonnell, S.; Swartwout, R.; Citorik, R.J.; et al. An Ingestible Bacterial-Electronic System to Monitor Gastrointestinal Health. Science 2018, 360, 915–918. [Google Scholar] [CrossRef]
- Wang, J.; Coleman, D.C.; Kanter, J.; Ummer, B.; Siminerio, L. Connecting Smartphone and Wearable Fitness Tracker Data with a Nationally Used Electronic Health Record System for Diabetes Education to Facilitate Behavioral Goal Monitoring in Diabetes Care: Protocol for a Pragmatic Multi-Site Randomized Trial. JMIR Res. Protoc. 2018, 7, e10009. [Google Scholar] [CrossRef]
- Eades, M.T.; Tsanas, A.; Juraschek, S.P.; Kramer, D.B.; Gervino, E.V.; Mukamal, K.J. Smartphone-Recorded Physical Activity for Estimating Cardiorespiratory Fitness. Sci. Rep. 2021, 11, 14851. [Google Scholar] [CrossRef]
- Seifert, A.; Schlomann, A.; Rietz, C.; Schelling, H.R. The Use of Mobile Devices for Physical Activity Tracking in Older Adults’ Everyday Life. Digit. Health 2017, 3, 205520761774008. [Google Scholar] [CrossRef]
- De Ridder, B.; Van Rompaey, B.; Kampen, J.K.; Haine, S.; Dilles, T. Smartphone Apps Using Photoplethysmography for Heart Rate Monitoring: Meta-Analysis. JMIR Cardio 2018, 2, e4. [Google Scholar] [CrossRef]
- Pipitprapat, W.; Harnchoowong, S.; Suchonwanit, P.; Sriphrapradang, C. The Validation of Smartphone Applications for Heart Rate Measurement. Ann. Med. 2018, 50, 721–727. [Google Scholar] [CrossRef]
- Chan, A.H.Y.; Pleasants, R.A.; Dhand, R.; Tilley, S.L.; Schworer, S.A.; Costello, R.W.; Merchant, R. Digital Inhalers for Asthma or Chronic Obstructive Pulmonary Disease: A Scientific Perspective. Pulm. Ther. 2021, 7, 345–376. [Google Scholar] [CrossRef]
- Zabczyk, C.; Blakey, J.D. The Effect of Connected Smart Inhalers on Medication Adherence. Front. Med. Technol. 2021, 3, 657321. [Google Scholar] [CrossRef]
- Le, M.-P.T.; Voigt, L.; Nathanson, R.; Maw, A.M.; Johnson, G.; Dancel, R.; Mathews, B.; Moreira, A.; Sauthoff, H.; Gelabert, C.; et al. Comparison of Four Handheld Point-Of-Care Ultrasound Devices by Expert Users. Ultrasound J. 2022, 14, 27. [Google Scholar] [CrossRef]
- Kwon, J.M.; Kim, K.H.; Jeon, K.H.; Lee, S.Y.; Park, J.; Oh, B.H. Artificial Intelligence Algorithm for Predicting Cardiac Arrest using Electrocardiography. Scand. J. Trauma Resusc. Emerg. Med. 2020, 28, 98. [Google Scholar] [CrossRef]
- Biondi, A.; Santoro, V.; Viana, P.F.; Laiou, P.; Pal, D.K.; Bruno, E.; Richardson, M.P. Noninvasive mobile EEG as a Tool for Seizure Monitoring and Management: A Systematic Review. Epilepsia 2022, 63, 1041–1063. [Google Scholar] [CrossRef]
- Kviesis-Kipge, E.; Rubins, U. Portable Remote Photoplethysmography Device for Monitoring of Blood Volume Changes with High Temporal Resolution. In Proceedings of the 2016 15th Biennial Baltic Electronics Conference (BEC), Tallinn, Estonia, 3–5 October 2016. [Google Scholar] [CrossRef]
- Zhou, J.; Li, X.; Wang, X.; Yu, N.; Wang, W. Accuracy of Portable Spirometers in The Diagnosis of Chronic Obstructive Pulmonary Disease a Meta-Analysis. npj Prim. Care Respir. Med. 2022, 32, 15. [Google Scholar] [CrossRef]
- Papadea, C.; Foster, J.; Grant, S.; Ballard, S.A.; Cate IV, J.C.; Michael Southgate, W.; Purohit, D.M. Evaluation of the I-STAT Portable Clinical Analyzer for Point-Of-Care Blood Testing in The Intensive Care Units of a University Children’s Hospital. Ann. Clin. Lab. Sci. 2002, 32, 231–243. [Google Scholar]
- Schrading, W.A.; McCafferty, B.; Grove, J.; Page, D.B. Portable, Consumer-Grade Pulse Oximeters Are Accurate for Home and Medical Use: Implications for Use in the COVID-19 Pandemic and Other Resource-Limited Environments. J. Am. Coll. Emerg. Physicians Open 2020, 1, 1450–1458. [Google Scholar] [CrossRef]
- Zijp, T.R.; Touw, D.J.; van Boven, J.F.M. User Acceptability and Technical Robustness Evaluation of a Novel Smart Pill Bottle Prototype Designed to Support Medication Adherence. Patient Prefer. Adherence 2020, 14, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Mubeen, I.; Ullah, N.; Shah, S.S.U.D.; Khan, B.A.; Zahoor, M.; Ullah, R.; Khan, F.A.; Sultan, M.A. Modern Diagnostic Imaging Technique Applications and Risk Factors in the Medical Field: A Review. BioMed Res. Int. 2022, 2022, 5164970. [Google Scholar] [CrossRef] [PubMed]
- Bi, W.L.; Hosny, A.; Schabath, M.B.; Giger, M.L.; Birkbak, N.J.; Mehrtash, A.; Allison, T.; Arnaout, O.; Abbosh, C.; Dunn, I.F.; et al. Artificial Intelligence in Cancer Imaging: Clinical Challenges and Applications. CA Cancer J. Clin. 2019, 69, 127–157. [Google Scholar] [CrossRef]
- Gulum, M.A.; Trombley, C.M.; Kantardzic, M. A Review of Explainable Deep Learning Cancer Detection Models in Medical Imaging. Appl. Sci. 2021, 11, 4573. [Google Scholar] [CrossRef]
- Rana, M.; Bhushan, M. Machine Learning and Deep Learning Approach for Medical Image Analysis: Diagnosis to Detection. Multimed. Tools Appl. 2022, 82, 26731–26769. [Google Scholar] [CrossRef] [PubMed]
- Iyawa, G.E.; Herselman, M.; Botha, A. Digital Health Innovation Ecosystems: From Systematic Literature Review to Conceptual Framework. Procedia Comput. Sci. 2016, 100, 244–252. [Google Scholar] [CrossRef]
- Vadillo Moreno, L.; Martín Ruiz, M.L.; Malagón Hernández, J.; Valero Duboy, M.Á.; Lindén, M. The Role of Smart Homes in Intelligent Homecare and Healthcare Environments. Ambient Assist. Living Enhanc. Living Environ. 2017, 345–394. [Google Scholar] [CrossRef]
- Chang, K.D.; Raheem, A.A.; Rha, K.H. Novel Robotic Systems and Future Directions. Indian J. Urol. 2018, 34, 110–114. [Google Scholar] [CrossRef]
- St Mart, J.P.; Goh, E.L. The Current State of Robotics in Total Knee Arthroplasty. EFORT Open Rev. 2021, 6, 270–279. [Google Scholar] [CrossRef]
- Acker, G.; Hashemi, S.M.; Fuellhase, J.; Kluge, A.; Conti, A.; Kufeld, M.; Kreimeier, A.; Loebel, F.; Kord, M.; Sladek, D.; et al. Efficacy and safety of CyberKnife Radiosurgery in Elderly Patients with Brain Metastases: A Retrospective Clinical Evaluation. Radiat. Oncol. 2020, 15, 225. [Google Scholar] [CrossRef]
- Ayano, Y.M.; Schwenker, F.; Dufera, B.D.; Debelee, T.G. Interpretable Machine Learning Techniques in ECG-Based Heart Disease Classification: A Systematic Review. Diagnostics 2022, 13, 111. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.A.; Ali, W.; Abdullah, T.A.A.; Malebary, S.J. Classifying Cardiac Arrhythmia from ECG Signal Using 1D CNN Deep Learning Model. Mathematics 2023, 11, 562. [Google Scholar] [CrossRef]
- El Idrissi, T.; Idri, A. Deep Learning for Blood Glucose Prediction: CNN vs. LSTM. In Computational Science and Its Applications—ICCSA 2020. ICCSA 2020; Gervasi, O., Murgante, B., Misra, S., Garau, C., Blečić, I., Taniar, D., Apduhan, B.O., Rocha, A.M.A.C., Tarantino, E., Torre, C.M., et al., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2020; Volume 12250, pp. 379–393. [Google Scholar] [CrossRef]
- Sun, Q.; Jankovic, M.V.; Bally, L.; Mougiakakou, S.G. Predicting Blood Glucose with an LSTM and Bi-LSTM Based Deep Neural Network. In Proceedings of the 2018 14th Symposium on Neural Networks and applications (NEUREL), Belgrade, Serbia, 20–21 November 2018; pp. 1–5. [Google Scholar] [CrossRef]
- Abineza, C.; Balas, V.E.; Nsengiyumva, P. A Machine-Learning-based Prediction Method for Easy COPD Classification based on Pulse Oximetry clinical Use. J. Intell. Fuzzy Syst. 2022, 43, 1683–1695. [Google Scholar] [CrossRef]
- Rohmetra, H.; Raghunath, N.; Narang, P.; Chamola, V.; Guizani, M.; Lakkaniga, N. AI-Enabled Remote Monitoring of Vital Signs for COVID-19: Methods, Prospects and Challenges. Computing 2021, 105, 783–809. [Google Scholar] [CrossRef]
- Lai, Z.; Vadlaputi, P.; Tancredi, D.J.; Garg, M.; Koppel, R.I.; Goodman, M.; Hogan, W.; Cresalia, N.; Juergensen, S.; Manalo, E.; et al. Enhanced Critical Congenital Cardiac Disease Screening by Combining Interpretable Machine Learning Algorithms. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2021, 2021, 1403–1406. [Google Scholar] [CrossRef] [PubMed]
- Rajaguru, H.; Prabhakar, S.K. Sparse PCA and Soft Decision Tree Classifiers for Epilepsy Classification from EEG Signals. In Proceedings of the 2017 International conference of Electronics, Communication and Aerospace Technology (ICECA), Coimbatore, India, 20–22 April 2017; Volume 1, pp. 581–584. [Google Scholar] [CrossRef]
- Sharma, A.; Rai, J.K.; Tewari, R.P. Epileptic Seizure Anticipation and Localisation of Epileptogenic Region Using EEG Signals. J. Med. Eng. Technol. 2018, 42, 203–216. [Google Scholar] [CrossRef]
- Alturki, F.A.; AlSharabi, K.; Abdurraqeeb, A.M.; Aljalal, M. EEG Signal Analysis for Diagnosing Neurological Disorders Using Discrete Wavelet Transform and Intelligent Techniques. Sensors 2020, 20, 2505. [Google Scholar] [CrossRef]
- Tawhid, M.N.A.; Siuly, S.; Wang, H. Diagnosis of Autism Spectrum Disorder from EEG using a Time–Frequency Spectrogram Image-based Approach. Electron. Lett. 2020, 56, 1372–1375. [Google Scholar] [CrossRef]
- Maray, N.; Ngu, A.H.; Ni, J.; Debnath, M.; Wang, L. Transfer Learning on Small Datasets for Improved Fall Detection. Sensors 2023, 23, 1105. [Google Scholar] [CrossRef]
- Butt, A.; Narejo, S.; Anjum, M.R.; Yonus, M.U.; Memon, M.; Samejo, A.A. Fall Detection Using LSTM and Transfer Learning. Wirel. Pers. Commun. 2022, 126, 1733–1750. [Google Scholar] [CrossRef]
- Kaisti, M.; Panula, T.; Leppänen, J.; Punkkinen, R.; Jafari Tadi, M.; Vasankari, T.; Jaakkola, S.; Kiviniemi, T.; Airaksinen, J.; Kostiainen, P.; et al. Clinical Assessment of a Non-Invasive Wearable MEMS Pressure Sensor Array for Monitoring of Arterial Pulse Waveform, Heart Rate and Detection of Atrial Fibrillation. npj Digit. Med. 2019, 2, 39. [Google Scholar] [CrossRef] [PubMed]
- El Asnaoui, K.; Chawki, Y. Using X-Ray Images and Deep Learning for Automated Detection of Coronavirus Disease. J. Biomol. Struct. Dyn. 2021, 39, 3615–3626. [Google Scholar] [CrossRef] [PubMed]
- Erdaw, Y.; Tachbele, E. Machine Learning Model Applied on Chest X-ray Images Enables Automatic Detection of COVID-19 Cases with High Accuracy. Int. J. Gen. Med. 2021, 14, 4923–4931. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Kodera, S.; Shinohara, H.; Ieki, H.; Yamaguchi, T.; Higashikuni, Y.; Kiyosue, A.; Ito, K.; Ando, J.; Takimoto, E.; et al. Diagnosing Heart Failure from Chest X-Ray Images Using Deep Learning. Int. Heart J. 2020, 61, 781–786. [Google Scholar] [CrossRef] [PubMed]
- Leiner, T.; Rueckert, D.; Suinesiaputra, A.; Baeßler, B.; Nezafat, R.; Išgum, I.; Young, A.A. Machine learning in cardiovascular magnetic resonance: Basic concepts and applications. J. Cardiovasc. Magn. Reson. 2019, 21, 61. [Google Scholar] [CrossRef] [PubMed]
- Chato, L.; Latifi, S. Machine Learning and Radiomic Features to Predict Overall Survival Time for Glioblastoma Patients. J. Pers. Med. 2021, 11, 1336. [Google Scholar] [CrossRef]
- Li, H.; Lee, C.H.; Chia, D.; Lin, Z.; Huang, W.; Tan, C.H. Machine Learning in Prostate MRI for Prostate Cancer: Current Status and Future Opportunities. Diagnostics 2022, 12, 289. [Google Scholar] [CrossRef]
- Siouras, A.; Moustakidis, S.; Giannakidis, A.; Chalatsis, G.; Liampas, I.; Vlychou, M.; Hantes, M.; Tasoulis, S.; Tsaopoulos, D. Knee Injury Detection Using Deep Learning on MRI Studies: A Systematic Review. Diagnostics 2022, 12, 537. [Google Scholar] [CrossRef]
- Said, Y.; Alsheikhy, A.A.; Shawly, T.; Lahza, H. Medical Images Segmentation for Lung Cancer Diagnosis Based on Deep Learning Architectures. Diagnostics 2023, 13, 546. [Google Scholar] [CrossRef]
- Sreenivasu, S.V.N.; Gomathi, S.; Kumar, M.J.; Prathap, L.; Madduri, A.; Almutairi, K.M.A.; Alonazi, W.B.; Kali, D.; Jayadhas, S.A. Dense Convolutional Neural Network for Detection of Cancer from CT Images. BioMed Res. Int. 2022, 2022, 1293548. [Google Scholar] [CrossRef]
- Dai, X.; Huang, L.; Qian, Y.; Xia, S.; Chong, W.; Liu, J.; Di Ieva, A.; Hou, X.; Ou, C. Deep Learning for Automated Cerebral Aneurysm Detection on Computed Tomography Images. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Abraham, G.K.; Bhaskaran, P.; Jayanthi, V.S. Lung Nodule Classification in CT Images Using Convolutional Neural Network. In Proceedings of the 2019 9th International Conference on Advances in Computing and Communication (ICACC), Kochi, India, 6–8 November 2019; pp. 199–203. [Google Scholar] [CrossRef]
- Vidhya, V.; Gudigar, A.; Raghavendra, U.; Hegde, A.; Menon, G.R.; Molinari, F.; Ciaccio, E.J.; Acharya, U.R. Automated Detection and Screening of Traumatic Brain Injury (TBI) Using Computed Tomography Images: A Comprehensive Review and Future Perspectives. Int. J. Environ. Res. Public Health 2021, 18, 6499. [Google Scholar] [CrossRef]
- Shinohara, I.; Inui, A.; Mifune, Y.; Nishimoto, H.; Yamaura, K.; Mukohara, S.; Yoshikawa, T.; Kato, T.; Furukawa, T.; Hoshino, Y.; et al. Using Deep Learning for Ultrasound Images to Diagnose Carpal Tunnel Syndrome with High Accuracy. Ultrasound Med. Biol. 2022, 48, 2052–2059. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, U.F.; Almekkawy, M. Automated Detection of Liver Steatosis in Ultrasound Images Using Convolutional Neural Networks. In Proceedings of the 2021 IEEE International Ultrasonics Symposium (IUS) 2021, Xi’an, China, 11–16 September 2021; pp. 1–4. [Google Scholar] [CrossRef]
- Xu, S.S.-D.; Chang, C.-C.; Su, C.-T.; Phu, P.Q. Classification of Liver Diseases Based on Ultrasound Image Texture Features. Appl. Sci. 2019, 9, 342. [Google Scholar] [CrossRef]
- Lv, Y.; Huang, Z. Account of Deep Learning-Based Ultrasonic Image Feature in the Diagnosis of Severe Sepsis Complicated with Acute Kidney Injury. Comput. Math. Methods Med. 2022, 2022, 8158634. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.M.; Adamali, H.; Molyneaux, P.L.; Lukey, P.T.; Marshall, R.P.; Renzoni, E.A.; Wells, A.U.; Maher, T.M. Daily Home Spirometry: An Effective Tool for Detecting Progression in Idiopathic Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2016, 194, 989–997. [Google Scholar] [CrossRef]
- Jung, T.; Vij, N. Early Diagnosis and Real-Time Monitoring of Regional Lung Function Changes to Prevent Chronic Obstructive Pulmonary Disease Progression to Severe Emphysema. J. Clin. Med. 2021, 10, 5811. [Google Scholar] [CrossRef]
- Islam, T.T.; Ahmed, M.S.; Hassanuzzaman, M.; Bin Amir, S.A.; Rahman, T. Blood Glucose Level Regression for Smartphone PPG Signals Using Machine Learning. Appl. Sci. 2021, 11, 618. [Google Scholar] [CrossRef]
- Susana, E.; Ramli, K.; Murfi, H.; Apriantoro, N.H. Non-Invasive Classification of Blood Glucose Level for Early Detection Diabetes Based on Photoplethysmography Signal. Information 2022, 13, 59. [Google Scholar] [CrossRef]
- Celka, P.; Charlton, P.H.; Farukh, B.; Chowienczyk, P.; Alastruey, J. Influence of Mental Stress on The Pulse Wave Features of Photoplethysmograms. Healthc. Technol. Lett. 2019, 7, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Přibil, J.; Přibilová, A.; Frollo, I. Stress Level Detection and Evaluation from Phonation and PPG Signals Recorded in an Open-Air MRI Device. Appl. Sci. 2021, 11, 11748. [Google Scholar] [CrossRef]
- Fan, J.; Sun, C.; Long, M.; Chen, C.; Chen, W. EOGNET: A Novel Deep Learning Model for Sleep Stage Classification Based on Single-Channel EOG Signal. Front. Neurosci. 2021, 15, 573194. [Google Scholar] [CrossRef] [PubMed]
- Weinhouse, G.L.; Kimchi, E.; Watson, P.; Devlin, J.W. Sleep Assessment in Critically Ill Adults: Established Methods and Emerging Strategies. Crit. Care Explor. 2022, 4, e0628. [Google Scholar] [CrossRef]
- Tag, B.; Vargo, A.W.; Gupta, A.; Chernyshov, G.; Kunze, K.; Dingler, T. Continuous Alertness Assessments. In Proceedings of the 2019 CHI Conference on Human Factors in Computing Systems, Glasgow, UK, 4–9 May 2019. [Google Scholar] [CrossRef]
- Hausfater, P.; Zhao, Y.; Defrenne, S.; Bonnet, P.; Riou, B. Cutaneous Infrared Thermometry for Detecting Febrile Patients. Emerg. Infect. Dis. 2008, 14, 1255–1258. [Google Scholar] [CrossRef] [PubMed]
- Sessler, D.I. Temperature Monitoring and Perioperative Thermoregulation. Anesthesiology 2008, 109, 318–338. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, G.J.; Caprioli, J. Optical Coherence Tomography to Detect and Manage Retinal Disease and Glaucoma. Am. J. Ophthalmol. 2004, 137, 156–169. [Google Scholar] [CrossRef]
- Le, D.; Son, T.; Yao, X. Machine Learning in Optical Coherence Tomography Angiography. Exp. Biol. Med. 2021, 246, 2170–2183. [Google Scholar] [CrossRef]
- Ahmed, M. Video Capsule Endoscopy in Gastroenterology. Gastroenterol. Res. 2022, 15, 47–55. [Google Scholar] [CrossRef]
- Akpunonu, B.; Hummell, J.; Akpunonu, J.D.; Ud Din, S. Capsule Endoscopy in Gastrointestinal Disease: Evaluation, Diagnosis, And Treatment. Clevel. Clin. J. Med. 2022, 89, 200–211. [Google Scholar] [CrossRef]
- Berus, L.; Klancnik, S.; Brezocnik, M.; Ficko, M. Classifying Parkinson’s Disease Based on Acoustic Measures Using Artificial Neural Networks. Sensors 2018, 19, 16. [Google Scholar] [CrossRef] [PubMed]
- Lahmiri, S.; Dawson, D.A.; Shmuel, A. Performance of Machine Learning Methods in Diagnosing Parkinson’s Disease Based on Dysphonia Measures. Biomed. Eng. Lett. 2017, 8, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Reid, J.; Parmar, P.; Lund, T.; Aalto, D.K.; Jeffery, C.C. Development of a machine-learning based voice disorder screening tool. Am. J. Otolaryngol. 2022, 43, 103327. [Google Scholar] [CrossRef] [PubMed]
- Brunese, L.; Martinelli, F.; Mercaldo, F.; Santone, A. Deep Learning for Heart Disease Detection Through Cardiac Sounds. Procedia Comput. Sci. 2020, 176, 2202–2211. [Google Scholar] [CrossRef]
- Joudeh, I.O.; Cretu, A.-M.; Guimond, S.; Bouchard, S. Prediction of Emotional Measures via Electrodermal Activity (EDA) and Electrocardiogram (ECG). Eng. Proc. 2022, 27, 47. [Google Scholar] [CrossRef]
- Gorson, J.; Cunningham, K.; Worsley, M.; O’Rourke, E. Using Electrodermal Activity Measurements to Understand Student Emotions While Programming. In Proceedings of the 2022 ACM Conference on International Computing Education Research, 7–11 August 2022; Association for Computing Machinery: New York, NY, USA; Volume 1, pp. 105–119. [Google Scholar] [CrossRef]
- Rahma, O.N.; Putra, A.P.; Rahmatillah, A.; Putri, Y.S.K.A.; Fajriaty, N.D.; Ain, K.; Chai, R. Electrodermal Activity for Measuring Cognitive and Emotional Stress Level. J. Med. Signals Sens. 2022, 12, 155–162. [Google Scholar] [CrossRef]
- Ghasemi-Roudsari, S.; Al-Shimary, A.; Varcoe, B.; Byrom, R.; Kearney, L.; Kearney, M. A Portable Prototype Magnetometer to Differentiate Ischemic and Non-Ischemic Heart Disease in Patients with Chest Pain. PLoS ONE 2018, 13, e0191241. [Google Scholar] [CrossRef]
- Ha, S.; Choi, S. Convolutional Neural Networks for Human Activity Recognition Using Multiple Accelerometer and Gyroscope Sensors. In Proceedings of the 2016 International Joint Conference on Neural Networks (IJCNN), Vancouver, BC, Canada, 24–29 July 2016; pp. 381–388. [Google Scholar] [CrossRef]
- Mehrmohammadi, M.; Yoon, S.J.; Yeager, D.; Emelianov, S.Y. Photoacoustic Imaging for Cancer Detection and Staging. Curr. Mol. Imaging 2013, 2, 89–105. [Google Scholar] [CrossRef]
- Gharieb, R.R. Photoacoustic Imaging for Cancer Diagnosis: A Breast Tumor Example. In Photoacoustic Imaging; IntechOpen: London, UK, 2020. [Google Scholar] [CrossRef]
- Qiu, T.; Lan, Y.; Gao, W.; Zhou, M.; Liu, S.; Huang, W.; Zeng, S.; Pathak, J.L.; Yang, B.; Zhang, J. Photoacoustic Imaging as A Highly Efficient and Precise Imaging Strategy for The Evaluation of Brain Diseases. Quant. Imaging Med. Surg. 2021, 11, 2169–2186. [Google Scholar] [CrossRef]
- Wick, K.D.; Matthay, M.A.; Ware, L.B. Pulse Oximetry for The Diagnosis and Management of Acute Respiratory Distress Syndrome. Lancet Respir. Med. 2022, 10, 1086–1098. [Google Scholar] [CrossRef]
- Jawin, V.; Ang, H.-L.; Omar, A.; Thong, M.-K. Beyond Critical Congenital Heart Disease: Newborn Screening Using Pulse Oximetry for Neonatal Sepsis and Respiratory Diseases in a Middle-Income Country. PLoS ONE 2015, 10, e0137580. [Google Scholar] [CrossRef] [PubMed]
- Horry, M.J.; Chakraborty, S.; Paul, M.; Ulhaq, A.; Pradhan, B.; Saha, M.; Shukla, N. COVID-19 Detection Through Transfer Learning Using Multimodal Imaging Data. IEEE Access 2020, 8, 149808–149824. [Google Scholar] [CrossRef] [PubMed]
- Nian, R.; Liu, J.; Huang, B. A review on Reinforcement Learning: Introduction and Applications in Industrial Process Control. Comput. Chem. Eng. 2020, 139, 106886. [Google Scholar] [CrossRef]
- van Engelen, J.E.; Hoos, H.H. A Survey on Semi-Supervised Learning. Mach. Learn. 2019, 109, 373–440. [Google Scholar] [CrossRef]
- Weiss, K.; Khoshgoftaar, T.M.; Wang, D. A survey of transfer learning. J. Big Data 2016, 3, 1345–1459. [Google Scholar] [CrossRef]
- Zhang, A.; Xing, L.; Zou, J.; Wu, J.C. Shifting Machine Learning for Healthcare from Development to Deployment and From Models to Data. Nat. Biomed. Eng. 2022, 6, 1330–1345. [Google Scholar] [CrossRef] [PubMed]
- Sami, S.M.; Nasrabadi, N.M.; Rao, R. Deep Transductive Transfer Learning for Automatic Target Recognition. In Proceedings of the Automatic Target Recognition XXXIII, Orlando, FL, USA, 30 April–5 May 2023; SPIE: Bellingham, DC, USA, 2023; Volume 12521, pp. 31–40. [Google Scholar] [CrossRef]
- Kushibar, K.; Salem, M.; Valverde, S.; Rovira, À.; Salvi, J.; Oliver, A.; Lladó, X. Transductive Transfer Learning for Domain Adaptation in Brain Magnetic Resonance Image Segmentation. Front. Neurosci. 2021, 15, 608808. [Google Scholar] [CrossRef]
- Nishio, M.; Fujimoto, K.; Matsuo, H.; Muramatsu, C.; Sakamoto, R.; Fujita, H. Lung Cancer Segmentation with Transfer Learning: Usefulness of a Pretrained Model Constructed from an Artificial Dataset Generated Using a Generative Adversarial Network. Front. Artif. Intell. 2021, 4, 694815. [Google Scholar] [CrossRef]
- Tschandl, P.; Sinz, C.; Kittler, H. Domain-Specific Classification-Pretrained Fully Convolutional Network Encoders for Skin Lesion Segmentation. Comput. Biol. Med. 2019, 104, 111–116. [Google Scholar] [CrossRef]
- Oh, K.; Chung, Y.C.; Kim, K.W.; Kim, W.S.; Oh, I.S. Classification and Visualization of Alzheimer’s Disease using Volumetric Convolutional Neural Network and Transfer Learning. Sci. Rep. 2019, 9, 18150. [Google Scholar] [CrossRef]
- Raza, N.; Naseer, A.; Tamoor, M.; Zafar, K. Alzheimer Disease Classification through Transfer Learning Approach. Diagnostics 2023, 13, 801. [Google Scholar] [CrossRef]
- Martinez, M.; De Leon, P.L. Falls Risk Classification of Older Adults Using Deep Neural Networks and Transfer Learning. IEEE J. Biomed. Health Inform. 2020, 24, 144–150. [Google Scholar] [CrossRef] [PubMed]
- De Bois, M.; El Yacoubi, M.A.; Ammi, M. Adversarial Multi-Source Transfer Learning in Healthcare: Application to Glucose Prediction for Diabetic People. Comput. Methods Programs Biomed. 2021, 199, 105874. [Google Scholar] [CrossRef] [PubMed]
- Iman, M.; Arabnia, H.R.; Rasheed, K. A Review of Deep Transfer Learning and Recent Advancements. Technologies 2023, 11, 40. [Google Scholar] [CrossRef]
- Zhang, X.; Liang, Y.; Li, W.; Liu, C.; Gu, D.; Sun, W.; Miao, L. Development and Evaluation of Deep Learning for Screening Dental Caries from Oral Photographs. Oral Dis. 2020, 28, 173–181. [Google Scholar] [CrossRef]
- Koike, T.; Qian, K.; Kong, Q.; Plumbley, M.D.; Schuller, B.W.; Yamamoto, Y. Audio for Audio is Better? An Investigation on Transfer Learning Models for Heart Sound Classification. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 74–77. [Google Scholar] [CrossRef]
- Chakraborty, A.; Anitescu, C.; Zhuang, X.; Rabczuk, T. Domain Adaptation based Transfer Learning Approach for Solving PDEs on Complex Geometries. Eng. Comput. 2022, 38, 4569–4588. [Google Scholar] [CrossRef]
- Wang, Y.-X.; Ramanan, D.; Hebert, M. Growing a Brain: Fine-Tuning by Increasing Model Capacity. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 3029–3038. [Google Scholar] [CrossRef]
- Fu, Z.; Zhang, B.; He, X.; Li, Y.; Wang, H.; Huang, J. Emotion Recognition based on Multi-Modal Physiological Signals and Transfer Learning. Front. Neurosci. 2022, 16, 1000716. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, X.; Che, T.; Bao, G.; Li, S. Multi-Task Deep Learning for Medical Image Computing and Analysis: A Review. Comput. Biol. Med. 2023, 153, 106496. [Google Scholar] [CrossRef]
- Torres-Soto, J.; Ashley, E.A. Multi-Task Deep Learning for Cardiac Rhythm Detection in Wearable Devices. npj Digit. Med. 2020, 3, 116. [Google Scholar] [CrossRef]
- Martindale, C.F.; Christlein, V.; Klumpp, P.; Eskofier, B.M. Wearables-based Multi-Task Gait and Activity Segmentation Using Recurrent Neural Networks. Neurocomputing 2021, 432, 250–261. [Google Scholar] [CrossRef]
- Hong, Y.; Wei, B.; Han, Z.; Li, X.; Zheng, Y.; Li, S. MMCL-Net: Spinal Disease Diagnosis in Global Mode Using Progressive Multi-Task Joint Learning. Neurocomputing 2020, 399, 307–316. [Google Scholar] [CrossRef]
- Sun, X.; Panda, R.; Feris, R.; Saenko, K. Adashare: Learning What to Share for Efficient Deep Multi-Task Learning. Adv. Neural Inf. Process. Syst. 2020, 33, 8728–8740. [Google Scholar]
- Liu, S.; Johns, E.; Davison, A.J. End-to-End Multi-Task Learning with Attention. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 1871–1880. [Google Scholar]
- Gupta, S.; Punn, N.S.; Sonbhadra, S.K.; Agarwal, S. MAG-Net: Multi-Task Attention Guided Network for Brain Tumor Segmentation and Classification. In Big Data Analytics; Srirama, S.N., Lin, J.C.W., Bhatnagar, R., Agarwal, S., Reddy, P.K., Eds.; Springer: Berlin/Heidelberg, Germany, 2021; p. 13147. [Google Scholar] [CrossRef]
- Lin, B.; Ye, F.; Zhang, Y. A Closer Look at Loss Weighting in Multi-Task Learning. arXiv 2021, arXiv:2111.10603. [Google Scholar]
- Kendall, A.; Gal, Y.; Cipolla, R. Multi-Task Learning Using Uncertainty to Weigh Losses for Scene Geometry and Semantics. In Proceedings of the IEEE conference on computer vision and pattern recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7482–7491. [Google Scholar]
- Chen, Z.; Badrinarayanan, V.; Lee, C.Y.; Rabinovich, A. Gradnorm: Gradient Normalization for Adaptive Loss Balancing in Deep Multitask Networks. Int. Conf. Mach. Learn. 2018, 80, 794–803. [Google Scholar]
- Yu, T.; Kumar, S.; Gupta, A.; Levine, S.; Hausman, K.; Finn, C. Gradient Surgery for Multi-Task Learning. Adv. Neural Inf. Process. Syst. 2020, 33, 5824–5836. [Google Scholar]
- Liu, L.; Li, Y.; Kuang, Z.; Xue, J.; Chen, Y.; Yang, W.; Liao, Q.; Zhang, W.; Towards Impartial Multi-Task Learning. Int. Conf. Learn. 2021. Available online: https://openreview.net/forum?id=IMPnRXEWpvr (accessed on 7 December 2023).
- Snell, J.; Swersky, K.; Zemel, R. Prototypical Networks for Few-Shot Learning. In Proceedings of the 31st International Conference on Neural Information Processing Systems (NIPS’17), Long Beach, CA, USA, 4–9 December 2017; Curran Associates Inc.: New York, NY, USA, 2017; pp. 4080–4090. [Google Scholar]
- Chen, Y.; Liu, Z.; Xu, H.; Darrell, T.; Wang, X. Meta-Baseline: Exploring Simple Meta-Learning for Few-Shot Learning. In Proceedings of the 2021 IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 10–17 October 2021. [Google Scholar] [CrossRef]
- Wang, Y.; Yao, Q.; Kwok, J.T.; Ni, L.M. Generalizing from a Few Examples. ACM Comput. Surv. 2020, 53, 1–34. [Google Scholar] [CrossRef]
- Sheller, M.J.; Edwards, B.; Reina, G.A.; Martin, J.; Pati, S.; Kotrotsou, A.; Milchenko, M.; Xu, W.; Marcus, D.; Colen, R.R.; et al. Federated Learning in Medicine: Facilitating Multi-Institutional Collaborations Without Sharing Patient Data. Sci. Rep. 2020, 10, 12598. [Google Scholar] [CrossRef]
- Rieke, N.; Hancox, J.; Li, W.; Milletarì, F.; Roth, H.R.; Albarqouni, S.; Bakas, S.; Galtier, M.N.; Landman, B.A.; Maier-Hein, K.; et al. The Future of Digital Health with Federated Learning. npj Digit. Med. 2020, 3, 119. [Google Scholar] [CrossRef]
- Dash, T.; Chitlangia, S.; Ahuja, A.; Srinivasan, A. A Review of Some Techniques for Inclusion of Domain-Knowledge into Deep Neural Networks. Sci. Rep. 2022, 12, 1040. [Google Scholar] [CrossRef]
- Williams, J.; Tadesse, A.; Sam, T.; Sun, H.; Montañez, G.D. Limits of Transfer Learning. In Machine Learning, Optimization, and Data Science; Springer: Cham, Switzerland, 2021. [Google Scholar] [CrossRef]


















| Sensor | Function | Application |
|---|---|---|
| Accelerometer | Measures the phone’s movement and orientation |
|
| GPS | Provides location information |
|
| Gyroscope | Measures the phone’s rotation to detect changes in position |
|
| Photoplethysmography (PPG) | Measures the heart rate |
|
| Photodiode sensor (ambient light sensor) | Measures the amount of light in the user’s environment |
|
| Infrared (IR) sensor (proximity sensor) | Detects the presence of nearby objects or surfaces, as well as the contactless monitoring of vital signs |
|
| Transfer Learning Method | Feature Extraction | Fine-Tuning | Domain Adaptation |
|---|---|---|---|
| Source and Target Domains | Similar | Similar or related | Related but not different |
| Source and target tasks | Similar/related/different | Similar or related | Similar or related |
| Model complexity | Low/moderate | Moderate/high | High |
| Features |
|
|
|
| Challenges |
|
|
|
| Transfer Learning Method | Multitask | Federated | Few-/One-/Zero-Shot |
|---|---|---|---|
| Source and Target Domains | Similar | Multiple distributed Similar | Similar/related/different |
| Source and target tasks | Multiple and related | Similar | Similar or related |
| Model complexity | High | High | High |
| Features |
|
|
|
| Challenges |
|
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chato, L.; Regentova, E. Survey of Transfer Learning Approaches in the Machine Learning of Digital Health Sensing Data. J. Pers. Med. 2023, 13, 1703. https://doi.org/10.3390/jpm13121703
Chato L, Regentova E. Survey of Transfer Learning Approaches in the Machine Learning of Digital Health Sensing Data. Journal of Personalized Medicine. 2023; 13(12):1703. https://doi.org/10.3390/jpm13121703
Chicago/Turabian StyleChato, Lina, and Emma Regentova. 2023. "Survey of Transfer Learning Approaches in the Machine Learning of Digital Health Sensing Data" Journal of Personalized Medicine 13, no. 12: 1703. https://doi.org/10.3390/jpm13121703
APA StyleChato, L., & Regentova, E. (2023). Survey of Transfer Learning Approaches in the Machine Learning of Digital Health Sensing Data. Journal of Personalized Medicine, 13(12), 1703. https://doi.org/10.3390/jpm13121703