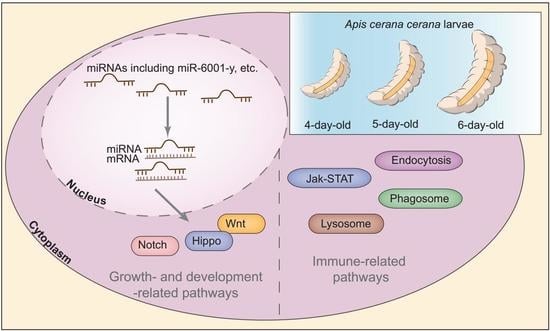
Expression Profile, Regulatory Network, and Putative Role of microRNAs in the Developmental Process of Asian Honey Bee Larval Guts
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Source of sRNA-seq Datasets
2.2. Identification and Analysis of miRNAs
2.3. Validation of miRNAs by Stem-Loop RT-PCR and Sanger Sequencing
2.4. Investigation of DEmiRNAs
2.5. RNA-seq Data Source
2.6. Prediction and Annotation of DEmiRNA-Targeted mRNAs
2.7. Construction and Investigation of DEmiRNA-DEmRNA Regulatory Networks
2.8. RT-qPCR Verification of DEmiRNAs
2.9. Statistical Analysis
3. Results
3.1. Analysis of miRNAs in the Larval Guts of A. c. cerana
3.2. Structural Characteristics of miRNAs in 4-, 5-, and 6-Day-Old Larval Guts
3.3. Expression Pattern of miRNAs during the Development Process of the A. c. cerana Larval Guts
3.4. Investigation and Annotation of DEmiRNA-Targeted mRNAs
3.5. DEmiRNA-mRNA Regulatory Network Engaged in the Development of A. c. cerana Larval Guts
3.6. DEmiRNA-mRNA Regulatory Network Involved in the Cellular and Humoral Immune of A. c. cerana Larval Guts
3.7. RT-qPCR Detection of DEmiRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ruttner, F. Biogeography and Taxonomy of Honeybees; Springer: Berlin, Germany; New York, NY, USA, 2013. [Google Scholar]
- Raymann, K.; Moran, N.A. The role of the gut microbiome in health and disease of adult honey bee workers. Curr. Opin. Insect Sci. 2018, 26, 97–104. [Google Scholar] [CrossRef]
- Giurfa, M.; Marcout, C.; Hilpert, P.; Thevenot, C.; Rugani, R. An insect brain organizes numbers on a left-to-right mental number line. Proc. Natl. Acad. Sci. USA 2022, 119, e2203584119. [Google Scholar] [CrossRef]
- Giurfa, M.; Zhang, S.; Jenett, A.; Menzel, R.; Srinivasan, M.V. The concepts of ‘sameness’ and ‘difference’ in an insect. Nature 2001, 410, 930–933. [Google Scholar] [CrossRef]
- Fan, X.; Zhang, W.; Zhang, K.; Zhang, J.; Long, Q.; Wu, Y.; Zhang, K.; Zhu, L.; Chen, D.; Guo, R. In-depth investigation of microRNA-mediated cross-kingdom regulation between Asian honey bee and microsporidian. Front. Microbiol. 2022, 13, 1003294. [Google Scholar] [CrossRef]
- Keller, A.; Brandel, A.; Becker, M.C.; Balles, R.; Abdelmohsen, U.R.; Ankenbrand, M.J.; Sickel, W. Wild bees and their nests host Paenibacillus bacteria with functional potential of avail. Microbiome 2018, 6, 229. [Google Scholar] [CrossRef] [PubMed]
- Watson, O.T.; Buchmann, G.; Young, P.; Lo, K.; Remnant, E.J.; Yagound, B.; Shambrook, M.; Hill, A.F.; Oldroyd, B.P.; Ashe, A. Abundant small RNAs in the reproductive tissues and eggs of the honey bee, Apis mellifera. BMC Genom. 2022, 23, 257. [Google Scholar] [CrossRef] [PubMed]
- Diao, Q.; Sun, L.; Zheng, H.; Zeng, Z.; Wang, S.; Xu, S.; Zheng, H.; Chen, Y.; Shi, Y.; Wang, Y.; et al. Genomic and transcriptomic analysis of the Asian honeybee Apis cerana provides novel insights into honeybee biology. Sci. Rep. 2018, 8, 822. [Google Scholar] [CrossRef]
- Winston, M.L. The Biology of the Honey Bee; Harvard University Press: Cambridge, UK, 1991. [Google Scholar]
- Hu, X.; Ke, L.; Wang, Z.; Zeng, Z. Dynamic transcriptome landscape of Asian domestic honeybee (Apis cerana) embryonic development revealed by high-quality RNA sequencing. BMC Dev. Biol. 2018, 18, 11. [Google Scholar] [CrossRef]
- Ai, H.; Yan, X.; Han, R. Occurrence and prevalence of seven bee viruses in Apis mellifera and Apis cerana apiaries in china. J. Invertebr. Pathol. 2012, 109, 160–164. [Google Scholar] [CrossRef]
- Zhang, Q.; Dou, W.; Taning, C.N.T.; Smagghe, G.; Wang, J.J. Regulatory roles of microRNAs in insect pests: Prospective targets for insect pest control. Curr. Opin. Biotechnol. 2021, 70, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, A.; Zhang, Y.; Xue, C.; Zhao, M.; Zhang, J. Activating pathway of three metabolic detoxification phases via down-regulated endogenous microRNAs, modulates triflumezopyrim tolerance in the small brown planthopper, Laodelphax striatellus (Fallén). Int. J. Biol. Macromol. 2022, 222, 2439–2451. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Du, B.; Xu, L.; Wang, H.; Wang, Y.; Lin, K.; He, G.; Kang, L. Glutamate-GABA imbalance mediated by miR-8-5p and its STTM regulates phase-related behavior of locusts. Proc. Natl. Acad. Sci. USA 2023, 120, e2215660120. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.J.; Zhu, F.; Liu, Y.J.; Li, Z.; Moural, T.W.; Liu, X.M.; Liu, X. MicroRNAs miR-14 and miR-2766 regulate tyrosine hydroxylase to control larval-pupal metamorphosis in Helicoverpa armigera. Pest Manag. Sci. 2022, 78, 3540–3550. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zou, H.; Lai, J.; Zhang, Z.; Sun, W. The miR-282-5p regulates larval moulting process by targeting chitinase 5 in Bombyx mori. Insect Mol. Biol. 2022, 31, 190–201. [Google Scholar] [CrossRef]
- Zhang, Q.; Dou, W.; Taning, C.N.T.; Yu, S.S.; Yuan, G.R.; Shang, F.; Smagghe, G.; Wang, J.J. miR-309a is a regulator of ovarian development in the oriental fruit fly Bactrocera dorsalis. PLoS Genet. 2022, 18, e1010411. [Google Scholar] [CrossRef]
- Freitas, F.C.; Pires, C.V.; Claudianos, C.; Cristino, A.S.; Simões, Z.L. MicroRNA-34 directly targets pair-rule genes and cytoskeleton component in the honey bee. Sci. Rep. 2017, 7, 40884. [Google Scholar] [CrossRef]
- Chen, X.; Ma, C.; Chen, C.; Lu, Q.; Shi, W.; Liu, Z.; Wang, H.; Guo, H. Integration of lncRNA-miRNA-mRNA reveals novel insights into oviposition regulation in honey bees. PeerJ 2017, 5, e3881. [Google Scholar] [CrossRef]
- Guo, X.; Su, S.; Skogerboe, G.; Dai, S.; Li, W.; Li, Z.; Liu, F.; Ni, R.; Guo, Y.; Chen, S.; et al. Recipe for a busy bee: MicroRNAs in honey bee caste determination. PLoS ONE 2013, 8, e81661. [Google Scholar] [CrossRef]
- Ma, L.; Liu, L.; Zhao, Y.; Yang, L.; Chen, C.; Li, Z.; Lu, Z. JNK pathway plays a key role in the immune system of the pea aphid and is regulated by microRNA-184. PLoS Pathog. 2020, 16, e1008627. [Google Scholar] [CrossRef]
- Huang, D.Y.; Xia, X.L.; Huang, R.; Li, S.; Yuan, D.W.; Liu, S.N. The steroid-induced microRNA let-7 regulates developmental growth by targeting cdc7 in the Drosophila fat body. Insect Sci. 2021, 28, 1621–1632. [Google Scholar] [CrossRef]
- Xiong, C.L.; Du, Y.; Chen, D.F.; Zheng, Y.Z.; Fu, Z.M.; Wang, H.P.; Geng, S.H.; Chen, H.Z.; Zhou, D.D.; Wu, S.Z.; et al. Bioinformatic prediction and analysis of miRNAs in the Apis mellifera ligustica larval gut. Chin. J. Appl. Entomol. 2018, 55, 1023–1033. (In Chinese) [Google Scholar]
- Guo, R.; Du, Y.; Xiong, C.L.; Zheng, Y.Z.; Fu, Z.M.; Xu, G.J.; Wang, H.P.; Chen, H.Z.; Geng, S.H.; Zhou, D.D.; et al. Differentially expressed microRNA and their regulation networks during the developmental process of Apis mellifera ligustica larval gut. Sci. Agric. Sin. 2018, 51, 4197–4209. (In Chinese) [Google Scholar]
- Chen, D.; Du, Y.; Chen, H.; Fan, Y.; Fan, X.; Zhu, Z.; Wang, J.; Xiong, C.; Zheng, Y.; Hou, C.; et al. Comparative identification of microRNAs in Apis cerana cerana workers’ midguts in responseto Nosema ceranae invasion. Insects 2019, 10, 258. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Zhao, H.; Shen, S.; Yang, S.; Yang, D.; Deng, S.; Hou, C. Identification of immune response to sacbrood virus infection in Apis cerana under natural condition. Front. Genet. 2020, 11, 587509. [Google Scholar] [CrossRef]
- Feng, R.R.; Fu, Z.M.; Du, Y.; Zhang, W.D.; Fan, X.X.; Wang, H.P.; Wan, J.Q.; Zhou, Z.Y.; Kang, Y.X.; Guo, R.; et al. Identification and analysis of micrornas in the larval gut of Apis cerana cerana. Sci. Agric. Sin. 2022, 55, 208–218. (In Chinese) [Google Scholar]
- Sharon, G.; Segal, D.; Ringo, J.M.; Hefetz, A.; Zilber-Rosenberg, I.; Rosenberg, E. Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2010, 107, 20051–20056. [Google Scholar] [CrossRef]
- Zheng, H.; Steele, M.I.; Leonard, S.P.; Motta, E.V.S.; Moran, N.A. Honey bees as models for gut microbiota research. Lab Anim. 2018, 47, 317–325. [Google Scholar] [CrossRef]
- Dosch, C.; Manigk, A.; Streicher, T.; Tehel, A.; Paxton, R.J.; Tragust, S. The gut microbiota can provide viral tolerance in the honey bee. Microorganisms 2021, 9, 871. [Google Scholar] [CrossRef]
- Pino, A.; Benkaddour, B.; Inturri, R.; Amico, P.; Vaccaro, S.C.; Russo, N.; Vaccalluzzo, A.; Agolino, G.; Caggia, C.; Miloud, H.; et al. Characterization of Bifidobacterium asteroides Isolates. Microorganisms 2022, 10, 655. [Google Scholar] [CrossRef]
- Long, Q.; Sun, M.H.; Fan, X.X.; Cai, Z.B.; Zhang, K.Y.; Wang, S.Y.; Zhang, J.X.; Gu, X.Y.; Song, Y.X.; Chen, D.F.; et al. First identification and investigation of piRNAs in the larval gut of the Asian honeybee, Apis cerana. Insects 2022, 14, 16. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.J.; Long, Q.; Fan, X.X.; Ye, Y.P.; Zhang, K.Y.; Zhang, J.X.; Zhao, H.D.; Yao, Y.T.; Fu, Z.M.; Chen, D.F.; et al. Transcriptome-wide characterization of piRNAs during the developmental process of european honey-bee larval guts. Genes 2022, 13, 1879. [Google Scholar] [CrossRef]
- Xu, X.J.; Guo, R.; Luo, Q.; Xiong, C.L.; Liang, Q.; Zhang, C.L.; Zheng, Y.Z.; Zhang, Z.N.; Huang, Z.J.; Zhang, L.; et al. De novo transcriptome assembly for Apis cerana cerana larval gut and identification of SSR molecular markers. Sci. Agric. Sin. 2017, 50, 1157–1167. [Google Scholar]
- Singh, V.K.; Mangalam, A.K.; Dwivedi, S.; Naik, S. Primer premier: Program for design of degenerate primers from a protein sequence. BioTechniques 1998, 24, 318–319. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Guo, Y.; Fan, X.; Zhao, H.; Zhang, Y.; Guo, S.; Jing, X.; Liu, Z.; Feng, P.; Liu, X.; et al. ame-miR-34 modulates the larval body weight and immune response of Apis mellifera workers to Ascosphara apis invasion. Int. J. Mol. Sci. 2023, 24, 1214. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Chen, D.; Diao, Q.; Xiong, C.; Zheng, Y.; Hou, C. Transcriptomic investigation of immune responses of the Apis cerana cerana larval gut infected by Ascosphaera apis. J. Invertebr. Pathol. 2019, 166, 107210. [Google Scholar] [CrossRef] [PubMed]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef] [PubMed]
- Krüger, J.; Rehmsmeier, M. RNAhybrid: MicroRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Anaysis of relative gene expression data using realtime quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Kim, M.C.; Lee, S.W.; Ryu, D.Y.; Cui, F.J.; Bhak, J.; Kim, Y. Identification and characterization of microRNAs in normal equine tissues by next generation sequencing. PLoS ONE 2014, 9, e93662. [Google Scholar] [CrossRef]
- Pan, Q.L.; Li, T.; He, Q.; Ma, Z.G.; Fan, X.D.; Zhang, X.Y.; Wang, Y.L.; Zhou, Z.Y.; Xu, J.S. Characterization of small RNAs in microsporidian Nosema bombycis. Acta Entomol. Sin. 2015, 58, 1213–1221. [Google Scholar] [CrossRef]
- Wang, X.; Wang, W.J.; Luo, Q.H.; Sun, G.H.; Feng, Y.W.; Ma, J.J.; Yang, J.M. Bioinformatics analysis of regulatory non-coding RNA in gonad of Crassostrea gigas. J. Fish. China 2020, 44, 723–734. [Google Scholar] [CrossRef]
- Mi, S.; Cai, T.; Hu, Y.; Chen, Y.; Hodges, E.; Ni, F.; Wu, L.; Li, S.; Zhou, H.; Long, C.; et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 2008, 133, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Shabalina, S.A.; Koonin, E.V. Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. 2008, 23, 578–587. [Google Scholar] [CrossRef]
- Eichhorn, S.W.; Guo, H.; McGeary, S.E.; Rodriguez-Mias, R.A.; Shin, C.; Baek, D.; Hsu, S.H.; Ghoshal, K.; Villén, J.; Bartel, D.P. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol. Cell 2014, 56, 104–115. [Google Scholar] [CrossRef]
- Steinhart, Z.; Angers, S. Wnt signaling in development and tissue homeostasis. Development 2018, 145, dev146589. [Google Scholar] [CrossRef]
- Choi, K.W. Upstream paths for Hippo signaling in Drosophila organ development. BMB Rep. 2018, 51, 134–142. [Google Scholar] [CrossRef]
- Cordero, J.B.; Sansom, O.J. Wnt signalling and its role in stem cell-driven intestinal regeneration and hyperplasia. Acta Physiol. 2012, 204, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Mah, A.T.; Yan, K.S.; Kuo, C.J. Wnt pathway regulation of intestinal stem cells. J. Physiol. 2016, 594, 4837–4847. [Google Scholar] [CrossRef]
- Russell, J.O.; Camargo, F.D. Hippo signalling in the liver: Role in development, regeneration and disease. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.; Woodgett, J.R. Glycogen synthase kinase 3: A kinase for all pathways? Curr. Top. Dev. Biol. 2017, 123, 277–302. [Google Scholar] [CrossRef]
- He, L.; Feng, H.; Yin, B.; Li, W.; Wang, X.; Umar, T.; Gao, H.; Zhou, N.; Qiu, C. Sodium new houttuyfonate induces apoptosis of breast cancer cells via ROS/PDK1/AKT/GSK3β Axis. Cancers 2023, 15, 1614. [Google Scholar] [CrossRef]
- Kanuka, H.; Kuranaga, E.; Takemoto, K.; Hiratou, T.; Okano, H.; Miura, M. Drosophila caspase transduces Shaggy/GSK-3β kinase activity in neural precursor development. EMBO J. 2005, 24, 3793–3806. [Google Scholar] [CrossRef]
- Wu, H.; Zhu, N.; Liu, J.; Ma, J.; Jiao, R. Shaggy regulates tissue growth through Hippo pathway in Drosophila. Sci. China Life Sci. 2022, 65, 2131–2144. [Google Scholar] [CrossRef] [PubMed]
- Karlovich, C.A.; Bonfini, L.; McCollam, L.; Rogge, R.D.; Daga, A.; Czech, M.P.; Banerjee, U. In vivo functional analysis of the Ras exchange factor son of sevenless. Science 1995, 268, 576–579. [Google Scholar] [CrossRef] [PubMed]
- Baltanás, F.C.; García-Navas, R.; Santos, E. SOS2 Comes to the Fore: Differential Functionalities in Physiology and Pathology. Int. J. Mol. Sci. 2021, 22, 6613. [Google Scholar] [CrossRef]
- Collins, D.H.; Mohorianu, I.; Beckers, M.; Moulton, V.; Dalmay, T.; Bourke, A.F. MicroRNAs associated with caste determination and differentiation in a primitively eusocial insect. Sci. Rep. 2017, 7, 45674. [Google Scholar] [CrossRef]
- Shi, Y.Y.; Zheng, H.J.; Pan, Q.Z.; Wang, Z.L.; Zeng, Z.J. Differentially expressed microRNAs between queen and worker larvae of the honey bee (Apis mellifera). Apidologie 2015, 46, 35–45. [Google Scholar] [CrossRef]
- Ashby, R.; Foret, S.; Searle, I.; Maleszka, R. MicroRNAs in honey bee caste determination. Sci. Rep. 2016, 6, 18794. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; An, S.; Cheng, P.; Zhang, K.; Gong, M.; Zhang, Z.; Zhang, R. Whole-transcriptome profiling across different developmental stages of Aedes albopictus (Diptera: Culicidae) provides insights into chitin-related non-coding RNA and competing endogenous RNA networks. Parasites Vectors 2023, 16, 33. [Google Scholar] [CrossRef] [PubMed]
- Mao, W.; Zeng, Q.; She, L.; Yuan, H.; Luo, Y.; Wang, R.; She, Y.; Wang, W.; Wang, C.; Pan, X. Wolbachia Utilizes lncRNAs to Activate the Anti-Dengue Toll Pathway and Balance Reactive Oxygen Species Stress in Aedes aegypti Through a Competitive Endogenous RNA Network. Front. Cell. Infect. Microbiol. 2022, 11, 823403. [Google Scholar] [CrossRef]
- Wang, Y.; Fu, Y.; Cheng, H.; Zhao, C.; Huang, Q.; Chang, M.; Qiu, W.; Shen, Y.; Li, D. lncR26319/miR-2834/EndophilinA axis regulates oogenesis of the silkworm, Bombyx mori. Insect Sci. 2023, 30, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Chen, H.Z.; Xiong, C.L.; Zheng, Y.Z.; Fu, Z.M.; Xu, G.J.; Du, Y.; Wang, H.P.; Geng, S.H.; Zhou, D.D.; et al. Analysis of differentially expressed circular RNAs and their regulation networks during the developmental process of Apis mellifera ligustica worker’s midgut. Sci. Agric. Sin. 2018, 51, 4575–4590. (In Chinese) [Google Scholar]
- Guo, R.; Geng, S.H.; Xiong, C.L.; Zheng, Y.Z.; Fu, Z.M.; Wang, H.P.; Du, Y.; Tong, X.Y.; Zhao, H.X.; Chen, D.F. Differential expression analysis of long non-coding RNAs during the developmental process of Apis mellifera ligustica worker’s midgut. Sci. Agric. Sin. 2018, 51, 3600–3613. (In Chinese) [Google Scholar]
- Fu, Z.M.; Gu, X.Y.; Hu, Y.; Zhao, H.D.; Zhu, Z.W.; Zhang, H.Y.; Ji, T.; Niu, Q.S.; Chen, D.F.; Guo, R. Lnc13164 regulates immune response of Apis cerana cerana larvae to Ascosphaera apis infection via ace-miR-4968-y. Acta Microbiol. Sin. 2023, 63, 1047–1059. [Google Scholar]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, X.; Zhang, W.; Guo, S.; Zhu, L.; Zhang, Y.; Zhao, H.; Gao, X.; Jiang, H.; Zhang, T.; Chen, D.; et al. Expression Profile, Regulatory Network, and Putative Role of microRNAs in the Developmental Process of Asian Honey Bee Larval Guts. Insects 2023, 14, 469. https://doi.org/10.3390/insects14050469
Fan X, Zhang W, Guo S, Zhu L, Zhang Y, Zhao H, Gao X, Jiang H, Zhang T, Chen D, et al. Expression Profile, Regulatory Network, and Putative Role of microRNAs in the Developmental Process of Asian Honey Bee Larval Guts. Insects. 2023; 14(5):469. https://doi.org/10.3390/insects14050469
Chicago/Turabian StyleFan, Xiaoxue, Wende Zhang, Sijia Guo, Leran Zhu, Yiqiong Zhang, Haodong Zhao, Xuze Gao, Haibin Jiang, Tianze Zhang, Dafu Chen, and et al. 2023. "Expression Profile, Regulatory Network, and Putative Role of microRNAs in the Developmental Process of Asian Honey Bee Larval Guts" Insects 14, no. 5: 469. https://doi.org/10.3390/insects14050469
APA StyleFan, X., Zhang, W., Guo, S., Zhu, L., Zhang, Y., Zhao, H., Gao, X., Jiang, H., Zhang, T., Chen, D., Guo, R., & Niu, Q. (2023). Expression Profile, Regulatory Network, and Putative Role of microRNAs in the Developmental Process of Asian Honey Bee Larval Guts. Insects, 14(5), 469. https://doi.org/10.3390/insects14050469