CD4+ T Cell Regulatory Network Underlies the Decrease in Th1 and the Increase in Anergic and Th17 Subsets in Severe COVID-19
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. CD4+ T Cell Regulatory Network
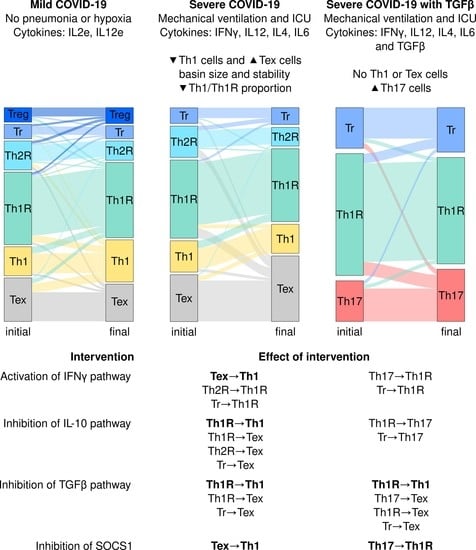
3.2. The Severity of COVID-19 Affects the Differentiation of CD4+ T Cells
3.3. Severe COVID-19 Decreases the Stability of Th1 Cells and Increases the Transitions towards Tex and Th17 Subsets
3.4. IFNG, TGF-, and SOCS1 Are Critical for the Immunomodulation of COVID-19
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mcgonagle, D.; Sharif, K.; Regan, A.O.; Bridgewood, C. The role of cytokines including IL-6 in COVID-19 induced pneumonia and MAS-like disease. Autoimmun. Rev. 2020, 19, 102537. [Google Scholar] [CrossRef]
- Ahmadpoor, P.; Rostaing, L. Why the immune system fails to mount an adaptive immune response to a COVID-19 infection. Transpl. Int. 2020, 33, 824–825. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Asquith, B.; Szydlo, R.; Tregoning, J.S.; Pollock, K.M. Peripheral T cell lymphopenia in COVID-19: Potential mechanisms and impact. Immunother. Adv. 2021, 1, ltab015. [Google Scholar] [CrossRef]
- Huang, I.; Pranata, R. Lymphopenia in severe coronavirus disease-2019 (COVID-19): Systematic review and meta-analysis. J. Intensive Care Vol. 2020, 8, 36. [Google Scholar] [CrossRef]
- Peng, X.; Ouyang, J.; Isnard, S.; Lin, J.; Fombuena, B.; Zhu, B.; Routy, J.P. Sharing CD4+ T Cell Loss: When COVID-19 and HIV Collide on Immune System. Front. Immunol. 2020, 11, 596631. [Google Scholar] [CrossRef]
- Choreño-Parra, J.A.; Jiménez-Álvarez, L.A.; Cruz-Lagunas, A.; Rodríguez-Reyna, T.S.; Ramírez-Martínez, G.; Sandoval-Vega, M.; Hernández-García, D.L.; Choreño-Parra, E.M.; Balderas-Martínez, Y.I.; Martinez-Sánchez, M.E.; et al. Clinical and Immunological Factors That Distinguish COVID-19 from Pandemic Influenza A(H1N1). Front. Immunol. 2021, 12, 1222. [Google Scholar] [CrossRef]
- Tjan, L.H.; Furukawa, K.; Nagano, T.; Kiriu, T.; Nishimura, M.; Arii, J.; Hino, Y.; Iwata, S.; Nishimura, Y.; Mori, Y. Early Differences in Cytokine Production by Severity of Coronavirus Disease 2019. J. Infect. Dis. 2021, 223, 1145–1149. [Google Scholar] [CrossRef]
- Han, H.; Ma, Q.; Li, C.; Liu, R.; Zhao, L.; Wang, W.; Zhang, P.; Liu, X.; Gao, G.; Liu, F.; et al. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg. Microbes Infect. 2020, 9, 1123–1130. [Google Scholar] [CrossRef]
- Wilk, A.J.; Rustagi, A.; Zhao, N.Q.; Roque, J.; Martínez-Colón, G.J.; McKechnie, J.L.; Ivison, G.T.; Ranganath, T.; Vergara, R.; Hollis, T.; et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020, 26, 1070–1076. [Google Scholar] [CrossRef]
- Noroozi, R.; Branicki, W.; Pyrc, K.; Łabaj, P.P.; Pospiech, E.; Taheri, M.; Ghafouri-Fard, S. Altered cytokine levels and immune responses in patients with SARS-CoV-2 infection and related conditions. Cytokine 2020, 133, 155143. [Google Scholar] [CrossRef]
- Ferreira-Gomes, M.; Kruglov, A.; Durek, P.; Heinrich, F.; Tizian, C.; Heinz, G.A.; Pascual-Reguant, A.; Du, W.; Mothes, R.; Fan, C.; et al. SARS-CoV-2 in severe COVID-19 induces a TGF-β-dominated chronic immune response that does not target itself. Nat. Commun. 2021, 12, 1961. [Google Scholar] [CrossRef]
- Mella, J.R.; Chiswick, E.L.; King, E.; Remick, D.G. Location, location, location: Cytokine concentrations are dependent on blood sampling site. Shock 2014, 42, 337–342. [Google Scholar] [CrossRef] [Green Version]
- Thurley, K.; Gerecht, D.; Friedmann, E.; Höfer, T. Three-dimensional gradients of cytokine signaling between T cells. PLoS Comput. Biol. 2015, 11, e1004206. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharyya, N.D.; Feng, C.G. Regulation of T Helper Cell Fate by TCR Signal Strength. Front. Immunol. 2020, 11, 624. [Google Scholar] [CrossRef]
- Gaud, G.; Lesourne, R.; Love, P.E. Regulatory mechanisms in T cell receptor signalling. Nat. Rev. Immunol. 2018, 18, 485–497. [Google Scholar] [CrossRef]
- Yamane, H.; Paul, W.E. Early signaling events that underlie fate decisions of naive CD4(+) T cells toward distinct T-helper cell subsets. Immunol. Rev. 2013, 252, 12–23. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Paul, W.E.W. Peripheral CD4+ T-cell differentiation regulated by networks of cytokines and transcription factors. Immunol. Rev. 2010, 238, 247–262. [Google Scholar] [CrossRef]
- Takada, K.; Jameson, S.C. Naive T cell homeostasis: From awareness of space to a sense of place. Nat. Rev. Immunol. 2009, 9, 823–832. [Google Scholar] [CrossRef]
- Ruterbusch, M.; Pruner, K.B.; Shehata, L.; Pepper, M. In Vivo CD4+ T Cell Differentiation and Function: Revisiting the Th1/Th2 Paradigm. Annu. Rev. Immunol. 2020, 38, 705–725. [Google Scholar] [CrossRef]
- Gagliani, N.; Huber, S. Basic aspects of T helper cell differentiation. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2017; Volume 1514, pp. 19–30. [Google Scholar] [CrossRef]
- Park, H.; Li, Z.; Yang, X.O.; Chang, S.H.; Nurieva, R.; Wang, Y.Y.H.; Hood, L.; Zhu, Z.; Tian, Q.; Dong, C.; et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 2005, 6, 1133–1141. [Google Scholar] [CrossRef] [Green Version]
- Vahedi, G.; Poholek, A.C.; Hand, T.W.; Laurence, A.; Kanno, Y.; O’Shea, J.J.; Hirahara, K.; Poholek, A.C.; Hand, T.W.; Shea, J.J.O.; et al. Helper T-cell identity and evolution of differential transcriptomes and epigenomes. Immunol. Rev. 2013, 252, 24–40. [Google Scholar] [CrossRef] [Green Version]
- Bilate, A.M.; Lafaille, J.J. Induced CD4(+)Foxp3(+) Regulatory T Cells in Immune Tolerance. Annu. Rev. Immunol. 2011, 30, 733–758. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Hong, J.; Sun, W.; Xu, G.; Li, N.; Chen, X.; Liu, A.; Xu, L.; Sun, B.; Zhang, J.Z. Role of IFN-γ in induction of Foxp3 and conversion of CD4 +CD25- T cells to CD4+ Tregs. J. Clin. Investig. 2006, 116, 2434–2441. [Google Scholar] [CrossRef] [Green Version]
- Daniel, V.; Wang, H.; Sadeghi, M.; Opelz, G. Interferon-gamma producing regulatory T cells as a diagnostic and therapeutic tool in organ transplantation. Int. Rev. Immunol. 2014, 33, 195–211. [Google Scholar] [CrossRef]
- Panduro, M.; Benoist, C.; Mathis, D. Tissue Tregs. Annu. Rev. Immunol. 2016, 34, 609–633. [Google Scholar] [CrossRef] [Green Version]
- Carrier, Y.; Yuan, J.; Kuchroo, V.K.V.K.; Weiner, H.L. Th3 Cells in Peripheral Tolerance. I. Induction of Foxp3-Positive Regulatory T Cells by Th3 Cells Derived from TGF-β T Cell-Transgenic Mice. J. Immunol. 2007, 178, 179–185. [Google Scholar] [CrossRef]
- Vieira, P.L.; Christensen, J.R.; Minaee, S.; O’Neill, E.J.; Barrat, F.J.; Boonstra, A.; Barthlott, T.; Stockinger, B.; Wraith, D.C.; O’Garra, A. IL-10-secreting regulatory T cells do not express Foxp3 but have comparable regulatory function to naturally occurring CD4+CD25+ regulatory T cells. J. Immunol. 2004, 172, 5986–5993. [Google Scholar] [CrossRef] [Green Version]
- Duan, W.; So, T.; Mehta, a.K.; Choi, H.; Croft, M. Inducible CD4+LAP+Foxp3- Regulatory T Cells Suppress Allergic Inflammation. J. Immunol. 2011, 187, 6499–6507. [Google Scholar] [CrossRef] [Green Version]
- Pace, L.; Rizzo, S.; Palombi, C.; Brombacher, F.; Doria, G. Cutting edge: IL-4-induced protection of CD4+CD25- Th cells from CD4+CD25+ regulatory T cell-mediated suppression. J. Immunol. 2006, 176, 3900–3904. [Google Scholar] [CrossRef]
- Duhen, T.; Duhen, R.; Lanzavecchia, A.; Sallusto, F.; Campbell, D.J. Functionally distinct subsets of human FOXP3 + Treg cells that phenotypically mirror effector Th cells. Blood 2012, 119, 4430–4440. [Google Scholar] [CrossRef] [Green Version]
- Fitzgerald, D.C.; Zhang, G.X.; El-Behi, M.; Fonseca-Kelly, Z.; Li, H.; Yu, S.; Saris, C.J.M.; Gran, B.; Ciric, B.; Rostami, A. Suppression of autoimmune inflammation of the central nervous system by interleukin 10 secreted by interleukin 27-stimulated T cells. Nat. Immunol. 2007, 8, 1372–1379. [Google Scholar] [CrossRef]
- Levine, A.G.; Medoza, A.; Hemmers, S.; Moltedo, B.; Niec, R.E.; Schizas, M.; Hoyos, B.E.; Putintseva, E.V.; Chaudhry, A.; Dikiy, S.; et al. Stability and function of regulatory T cells expressing the transcription factor T-bet. Nature 2017, 546, 421–425. [Google Scholar] [CrossRef] [Green Version]
- Yu, F.; Sharma, S.; Edwards, J.; Feigenbaum, L.; Zhu, J. Dynamic expression of transcription factors T-bet and GATA-3 by regulatory T cells maintains immunotolerance. Nat. Immunol. 2015, 16, 197–206. [Google Scholar] [CrossRef] [Green Version]
- Wohlfert, E.A.; Grainger, J.R.; Bouladoux, N.; Konkel, J.E.; Oldenhove, G.; Ribeiro, C.H.; Hall, J.A.; Yagi, R.; Naik, S.; Bhairavabhotla, R.; et al. GATA3 controls Foxp3+ regulatory T cell fate during inflammation in mice. J. Clin. Investig. 2011, 121, 4503–4515. [Google Scholar] [CrossRef] [Green Version]
- Yoshimura, A.; Suzuki, M.; Sakaguchi, R.; Hanada, T.; Yasukawa, H. SOCS, Inflammation, and Autoimmunity. Front. Immunol. 2012, 3, 20. [Google Scholar] [CrossRef] [Green Version]
- Knosp, C.A.; Johnston, J.A. Regulation of CD4+ T-cell polarization by suppressor of cytokine signalling proteins. Immunology 2012, 135, 101–111. [Google Scholar] [CrossRef]
- Blank, C.U.; Haining, W.N.; Held, W.; Hogan, P.G.; Kallies, A.; Lugli, E.; Lynn, R.C.; Philip, M.; Rao, A.; Restifo, N.P.; et al. Defining ‘T cell exhaustion’. Nat. Rev. Immunol. 2019, 19, 665–674. [Google Scholar] [CrossRef]
- Miggelbrink, A.M.; Jackson, J.D.; Lorrey, S.J.; Srinivasan, E.S.; Waibl-Polania, J.; Wilkinson, D.S.; Fecci, P.E. CD4 T-Cell Exhaustion: Does It Exist and What Are Its Roles in Cancer? Clin. Cancer Res. 2021, 27, 5742–5752. [Google Scholar] [CrossRef]
- Schwartz, R.H. T cell anergy. Annu. Rev. Immunol. 2003, 21, 305. [Google Scholar] [CrossRef]
- Kalekar, L.A.; Schmiel, S.E.; Nandiwada, S.L.; Lam, W.Y.; Barsness, L.O.; Zhang, N.; Stritesky, G.L.; Malhotra, D.; Pauken, K.E.; Linehan, J.L.; et al. CD4+ T cell anergy prevents autoimmunity and generates regulatory T cell precursors. Nat. Immunol. 2016, 17, 304–314. [Google Scholar] [CrossRef]
- Crespo, J.; Sun, H.; Welling, T.H.; Tian, Z.; Zou, W. T cell anergy, exhaustion, senescence, and stemness in the tumor microenvironment. Curr. Opin. Immunol. 2013, 25, 214–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fathi, N.; Rezaei, N. Lymphopenia in COVID-19: Therapeutic opportunities. Cell Biol. Int. 2020, 44, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Fang, X.; Tokuno, S.; Chung, U.; Chen, X.; Dai, X.; Liu, X.; Xu, F.; Wang, B.; Peng, P. A web visualization tool using T cell subsets as the predictor to evaluate COVID-19 patient’s severity. PLoS ONE 2020, 15, e0239695. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Jiang, L.; Li, X.; Lin, F.; Wang, Y.; Li, B.; Jiang, T.; An, W.; Liu, S.; Liu, H.; et al. Clinical and pathological investigation of patients with severe COVID-19. JCI Insight 2020, 5, e138070. [Google Scholar] [CrossRef]
- Kwiecień, I.; Rutkowska, E.; Kłos, K.; Wiȩsik-Szewczyk, E.; Jahnz-Różyk, K.; Rzepecki, P.; Chciałowski, A. Maturation of T and B Lymphocytes in the Assessment of the Immune Status in COVID-19 Patients. Cells 2020, 9, 2615. [Google Scholar] [CrossRef]
- Meckiff, B.J.; Ramírez-Suástegui, C.; Fajardo, V.; Chee, S.J.; Kusnadi, A.; Simon, H.; Eschweiler, S.; Grifoni, A.; Pelosi, E.; Weiskopf, D.; et al. Imbalance of Regulatory and Cytotoxic SARS-CoV-2-Reactive CD4+ T Cells in COVID-19. Cell 2020, 183, 1340–1353.e16. [Google Scholar] [CrossRef]
- Gao, M.; Liu, Y.; Guo, M.; Wang, Q.; Wang, Y.; Fan, J.; Shen, Y.; Hou, J.; Wan, Y.; Zhu, Z. Regulatory CD4+ and CD8+ T cells are negatively correlated with CD4+/CD8+ T cell ratios in patients acutely infected with SARS-CoV-2. J. Leukoc. Biol. 2021, 109, 91–97. [Google Scholar] [CrossRef]
- Neumann, J.; Prezzemolo, T.; Vanderbeke, L.; Roca, C.P.; Gerbaux, M.; Janssens, S.; Willemsen, M.; Burton, O.; Van Mol, P.; Van Herck, Y.; et al. Increased IL-10-producing regulatory T cells are characteristic of severe cases of COVID-19. Clin. Transl. Immunol. 2020, 9, e1204. [Google Scholar] [CrossRef]
- Galván-Peña, S.; Leon, J.; Chowdhary, K.; Michelson, D.A.; Vijaykumar, B.; Yang, L.; Magnuson, A.M.; Chen, F.; Manickas-Hill, Z.; Piechocka-Trocha, A.; et al. Profound Treg perturbations correlate with COVID-19 severity. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Rahimzadeh, M.; Naderi, N. Toward an understanding of regulatory T cells in COVID-19: A systematic review. J. Med. Virol. 2021, 93, 4167–4181. [Google Scholar] [CrossRef]
- Kauffman, S. Metabolic stability and epigenesis in randomly constructed genetic nets. J. Theor. Biol. 1969, 22, 437–467. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, L.; Thieffry, D.; Alvarez-Buylla, E.R. Genetic control of flower morphogenesis in Arabidopsis thaliana: A logical analysis. Bioinformatics 1999, 15, 593–606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albert, R.; Thakar, J. Boolean modeling: A logic-based dynamic approach for understanding signaling and regulatory networks and for making useful predictions. Wiley Interdiscip. Rev. Syst. Biol. Med. 2014, 6, 353–369. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, R.; Herrera, A.M.; Ramirez, J.; Qian, C.; Melton, D.W.; Shireman, P.K.; Jin, Y.F. Deriving a Boolean dynamics to reveal macrophage activation with in vitro temporal cytokine expression profiles. BMC Bioinform. 2019, 20, 725. [Google Scholar] [CrossRef] [Green Version]
- Liquitaya-Montiel, A.J.; Mendoza, L. Dynamical analysis of the regulatory network controlling natural killer cells differentiation. Front. Physiol. 2018, 9, 1029. [Google Scholar] [CrossRef]
- Avila-Ponce de León, U.; Vázquez-Jiménez, A.; Matadamas-Guzman, M.; Pelayo, R.; Resendis-Antonio, O. Transcriptional and Microenvironmental Landscape of Macrophage Transition in Cancer: A Boolean Analysis. Front. Immunol. 2021, 12, 2110. [Google Scholar] [CrossRef]
- Mendoza, L. A network model for the control of the differentiation process in Th cells. BioSystems 2006, 84, 101–114. [Google Scholar] [CrossRef]
- Carbo, A.; Hontecillas, R.; Kronsteiner, B.; Viladomiu, M.; Pedragosa, M.; Lu, P.; Philipson, C.W.; Hoops, S.; Marathe, M.; Eubank, S.; et al. Systems modeling of molecular mechanisms controlling cytokine-driven CD4+ T cell differentiation and phenotype plasticity. PLoS Comput. Biol. 2013, 9, e1003027. [Google Scholar] [CrossRef] [Green Version]
- Abou-Jaoudé, W.; Monteiro, P.T.; Naldi, A.; Grandclaudon, M.; Soumelis, V.; Chaouiya, C.; Thieffry, D. Model checking to assess T-helper cell plasticity. Front. Bioeng. Biotechnol. 2014, 2, 86. [Google Scholar] [CrossRef]
- Martinez-Sanchez, M.E.; Huerta, L.; Alvarez-Buylla, E.R.; Villarreal Luján, C. Role of Cytokine Combinations on CD4+ T Cell Differentiation, Partial Polarization, and Plasticity: Continuous Network Modeling Approach. Front. Physiol. 2018, 9, 877. [Google Scholar] [CrossRef]
- Martínez-Méndez, D.; Villarreal, C.; Mendoza, L.; Huerta, L. An Integrative Network Modeling Approach to T CD4 Cell Activation. Front. Physiol. 2020, 11, 380. [Google Scholar] [CrossRef] [PubMed]
- Müssel, C.; Hopfensitz, M.; Kestler, H.A.; Christoph, M.; Hopfensitz, M.; Kestler, H.A. BoolNet package vignette. Tutorial 2015, 49, 1–34. [Google Scholar]
- Zheng, Y.; Chaudhry, A.; Kas, A.; DeRoos, P.; Kim, J.M.; Chu, T.T.; Corcoran, L.; Treuting, P.; Klein, U.; Rudensky, A.Y.; et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature 2009, 458, 351–356. [Google Scholar] [CrossRef] [Green Version]
- Kalfaoglu, B.; Almeida-Santos, J.; Tye, C.A.; Satou, Y.; Ono, M. T-cell dysregulation in COVID-19. Biochem. Biophys. Res. Commun. 2021, 538, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Verma, N.D.; Hall, B.M.; Plain, K.M.; Robinson, C.M.; Boyd, R.; Tran, G.T.; Wang, C.; Bishop, G.A.; Hodgkinson, S.J. Interleukin-12 (IL-12p70) promotes induction of highly potent Th1-like CD4+CD25+ T regulatory cells that inhibit allograft rejection in unmodified recipients. Front. Immunol. 2014, 5, 190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- U.S. National Library of Medicine. NLM Drug Information Portal; U.S. National Library of Medicine: Bethesda, MD, USA, 2022.
- Jonuleit, H.; Schmitt, E.; Schuler, G.; Knop, J.; Enk, A.H. Induction of interleukin 10-producing, nonproliferating CD4(+) T cells with regulatory properties by repetitive stimulation with allogeneic immature human dendritic cells. J. Exp. Med. 2000, 192, 1213–1222. [Google Scholar] [CrossRef]
- Liu, Y.; Du, X.; Chen, J.; Jin, Y.; Peng, L.; Wang, H.H.; Luo, M.; Chen, L.; Zhao, Y. Neutrophil-to-lymphocyte ratio as an independent risk factor for mortality in hospitalized patients with COVID-19. J. Infect. 2020, 81, e6–e12. [Google Scholar] [CrossRef]
- Chiappelli, F. COVID-19 Immunopathology & Immunotherapy. Bioinformation 2020, 16, 219–222. [Google Scholar] [CrossRef]
- Sharma, J.; Larkin, J. Therapeutic implication of SOCS1 modulation in the treatment of autoimmunity and cancer. Front. Pharmacol. 2019, 10, 324. [Google Scholar] [CrossRef]
- Saez-Rodriguez, J.; Simeoni, L.; Lindquist, J.A.; Hemenway, R.; Bommhardt, U.; Arndt, B.; Haus, U.U.; Weismantel, R.; Gilles, E.D.; Klamt, S.; et al. A logical model provides insights into T cell receptor signaling. PLoS Comput. Biol. 2007, 3, 1580–1590. [Google Scholar] [CrossRef] [PubMed]
- Ross, S.H.; Cantrell, D.A. Signaling and Function of Interleukin-2 in T Lymphocytes. Annu. Rev. Immunol. 2018, 36, 411–433. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.K.; Gracias, D.T.; Croft, M. TNF activity and T cells. Cytokine 2018, 101, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.G.; Arvey, A.; Jin, W.; Rudensky, A.Y. Continuous requirement for the TCR in regulatory T cell function. Nat. Immunol. 2014, 15, 1070–1078. [Google Scholar] [CrossRef] [Green Version]
- Davignon, J.L.; Rauwel, B.; Degboé, Y.; Constantin, A.; Boyer, J.F.; Kruglov, A.; Cantagrel, A. Modulation of T-cell responses by anti-tumor necrosis factor treatments in rheumatoid arthritis: A review. Arthritis Res. Ther. 2018, 20, 229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubo, T.; Hatton, R.D.; Oliver, J.; Liu, X.; Elson, C.O.; Weaver, C.T. Regulatory T cell suppression and anergy are differentially regulated by proinflammatory cytokines produced by TLR-activated dendritic cells. J. Immunol. 2004, 173, 7249–7258. [Google Scholar] [CrossRef] [Green Version]
- Mendoza, L. A Virtual Culture of CD4+ T Lymphocytes. Bull. Math. Biol. 2013, 75, 1012–1029. [Google Scholar] [CrossRef]





| Environment | CoV-sev | CoV-TGFB | CoV-IL10 | |||
|---|---|---|---|---|---|---|
| Perturb Time | t = ∞ | t = 1 | t = ∞ | t = 1 | t = ∞ | t = 1 |
| IFNG = 1 | Tex → Th1 | Tex → Th1 | - | - | - | - |
| Th2R → Th1R, Tr → Th1R | Th2R → Th1R, Tr → Th1R | Th17 → Th1R, Tr → Th1R | Th17 → Th1R, Tr → Th1R | Th2R → Th1R, Tr → Th1R | Th2R → Th1R, Tr → Th1R | |
| IL10 = 0 | Th1R → Th1 | Th1R → Th1 | - | - | Th1R → Th1 | - |
| Th1R → Tex, Th2R → Tex, Tr → Tex | Th1R → Tex, Th2R → Tex, Tr → Tex | Th1R → Th17, Tr → Th17 | Th1R → Th17, Tr → Th17 | Th1R → Tex, Th2R → Tex, Tr → Tex | Th1R → Tr, Th2R → Tr, Tr → Th1R | |
| TGFB = 0 | Th1R → Th1 | Th1R → Th1 | Th1R → Th1 | - | - | - |
| Th1R → Tex, Tr → Tex | Th1R → Tex, Tr → Tex | Th17 → Tex, Th1R → Tex, Tr → Tex | Th1R → Th17, Tr → Th17 | - | - | |
| SOCS1 = 0 | Tex → Th1 | Tex → Th1 | - | - | - | - |
| - | - | Th17 → Th1R | Th17 → Th1R | - | - | |
| IL6 = 1 | - | Th1R → Th1 | - | - | - | - |
| Th1 → Tex, Th1R → Tex, Th1R → Tr, Th2R → Tr | Th1R → Tex, Th2R → Tex, Tr → Tex | Th1R → Tr | Th1R → Tr | Th1R → Tr, Th2R → Tr | Th1R → Tr, Th2R → Tr | |
| SOCS3 = 0 | - | Th1R → Th1 | - | - | - | - |
| Th1 → Tex, Th1R → Tex, Th1R → Tr, Th2R → Tr | Th1R → Tex, Th2R → Tex, Tr → Tex | Th1R → Th17, Th1R → Th17R, Tr → Th17R | Th1R → Tr | Th1R → Tr, Th2R → Tr | Th1R → Tr, Th2R → Tr | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martinez-Sánchez, M.E.; Choreño-Parra, J.A.; Álvarez-Buylla, E.R.; Zúñiga, J.; Balderas-Martínez, Y.I. CD4+ T Cell Regulatory Network Underlies the Decrease in Th1 and the Increase in Anergic and Th17 Subsets in Severe COVID-19. Pathogens 2023, 12, 18. https://doi.org/10.3390/pathogens12010018
Martinez-Sánchez ME, Choreño-Parra JA, Álvarez-Buylla ER, Zúñiga J, Balderas-Martínez YI. CD4+ T Cell Regulatory Network Underlies the Decrease in Th1 and the Increase in Anergic and Th17 Subsets in Severe COVID-19. Pathogens. 2023; 12(1):18. https://doi.org/10.3390/pathogens12010018
Chicago/Turabian StyleMartinez-Sánchez, Mariana Esther, José Alberto Choreño-Parra, Elena R. Álvarez-Buylla, Joaquín Zúñiga, and Yalbi Itzel Balderas-Martínez. 2023. "CD4+ T Cell Regulatory Network Underlies the Decrease in Th1 and the Increase in Anergic and Th17 Subsets in Severe COVID-19" Pathogens 12, no. 1: 18. https://doi.org/10.3390/pathogens12010018
APA StyleMartinez-Sánchez, M. E., Choreño-Parra, J. A., Álvarez-Buylla, E. R., Zúñiga, J., & Balderas-Martínez, Y. I. (2023). CD4+ T Cell Regulatory Network Underlies the Decrease in Th1 and the Increase in Anergic and Th17 Subsets in Severe COVID-19. Pathogens, 12(1), 18. https://doi.org/10.3390/pathogens12010018