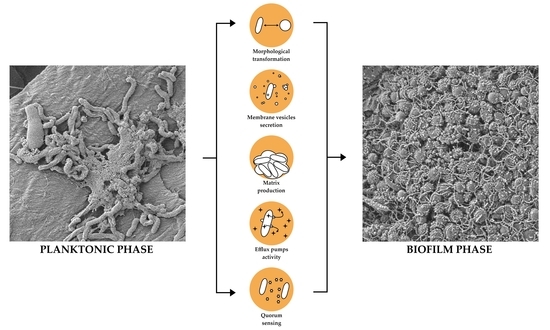
Biofilm Formation as a Complex Result of Virulence and Adaptive Responses of Helicobacter pylori
Abstract
:1. The Issue of Biofilm Formation in the Biomedical Sector
2. Characteristics of H. pylori Biofilms Produced in Laboratory Conditions
3. Transcriptomic and Proteomic Analysis of Biofilm H. pylori Forms
4. Phenotypic Variability as a Modulator of H. pylori Biofilm Structure
4.1. Coccoid Forms
4.2. Metabolic Activity and Matrix Production
4.3. Secretion of Outer Membrane Vesicles
4.4. Efflux Pump Activity
4.5. Quorum Sensing and a Role of Flagella in Biofilm Formation
5. Holistic Model Describing the Transition of H. pylori into the Biofilm Phase
6. Limitations and Challenges in H. pylori Biofilm Research
7. Conclusions
- Strong influence of virulence and adaptive responses (morphological transformation, membrane vesicles secretion, matrix production, efflux pump activity, and intermicrobial communication) on biofilm development and its structure.
- Higher expression of adhesins, lipopolysaccharide, flagella, components of T4SS systems, toxin-antitoxin systems, efflux pumps, enzymes regulating pH (e.g., urease) and responsible for obtaining alternative energy sources (e.g., hydrogenase), and proteins related to the cell wall rearrangement.
- Lower expression of factors involved in metabolism, translation, and quorum sensing related to the autoinducer-2 (AI-2) activity.
Author Contributions
Funding
Conflicts of Interest
References
- Langenheder, S.; Bulling, M.T.; Solan, M.; Prosser, J.I. Bacterial Biodiversity-Ecosystem Functioning Relations are Modified by Environmental Complexity. PLoS ONE 2010, 5, e10834. [Google Scholar] [CrossRef] [Green Version]
- Høiby, N.; Bjarnsholt, T.; Moser, C.; Bassi, G.L.; Coenye, T.; Donelli, G.; Hall-Stoodley, L.; Holá, V.; Imbert, C.; Kirketerp-Møller, K.; et al. ESCMID* Guideline for the Diagnosis and Treatment of Biofilm Infections 2014. Clin. Microbiol. Infect. 2015, 21, S1–S25. [Google Scholar] [CrossRef] [Green Version]
- Flemming, H.C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An Emergent Form of Bacterial Life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Lebeaux, D.; Ghigo, J.-M.; Beloin, C. Biofilm-Related Infections: Bridging the Gap between Clinical Management and Fundamental Aspects of Recalcitrance toward Antibiotics. Microbiol. Mol. Biol. Rev. 2014, 78, 510–543. [Google Scholar] [CrossRef] [Green Version]
- Vestby, L.K.; Grønseth, T.; Simm, R.; Nesse, L.L. Bacterial Biofilm and its Role in the Pathogenesis of Disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef] [Green Version]
- Paluch, E.; Rewak-Soroczyńska, J.; Jędrusik, I.; Mazurkiewicz, E.; Jermakow, K. Prevention of Biofilm Formation by Quorum Quenching. Appl. Microbiol. Biotechnol. 2020, 104, 1871–1881. [Google Scholar] [CrossRef] [Green Version]
- Høiby, N. A Short History of Microbial Biofilms and Biofilm Infections. APMIS 2017, 125, 272–275. [Google Scholar] [CrossRef] [Green Version]
- Ciofu, O.; Rojo-Molinero, E.; Macià, M.D.; Oliver, A. Antibiotic Treatment of Biofilm Infections. APMIS 2017, 125, 304–319. [Google Scholar] [CrossRef]
- Toh, J.W.T.; Wilson, R.B. Pathways of Gastric Carcinogenesis, Helicobacter pylori Virulence and Interactions with Antioxidant Systems, Vitamin C and Phytochemicals. Int. J. Mol. Sci. 2020, 21, 6451. [Google Scholar] [CrossRef]
- Alipour, M. Molecular Mechanism of Helicobacter pylori-Induced Gastric Cancer. J. Gastrointest. Cancer 2020, 14, 1–8. [Google Scholar] [CrossRef]
- Rhee, K.H.; Park, J.S.; Cho, M.J. Helicobacter pylori: Bacterial Strategy for Incipient Stage and Persistent Colonization in Human Gastric Niches. Yonsei Med. J. 2014, 55, 1453–1466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denic, M.; Touati, E.; De Reuse, H. Review: Pathogenesis of Helicobacter pylori Infection. Helicobacter 2020, 25, e12736. [Google Scholar] [CrossRef]
- Yamaoka, Y.; Graham, D.Y. Helicobacter pylori Virulence and Cancer Pathogenesis. Futur. Oncol. 2014, 10, 1487–1500. [Google Scholar] [CrossRef] [Green Version]
- Schulz, C.; Kupčinskas, J. Review—Helicobacter pylori and Non-Malignant Upper Gastro-Intestinal Diseases. Helicobacter 2020, 25, e12738. [Google Scholar] [CrossRef]
- Robinson, K. Helicobacter pylori-Mediated Protection against Extra-Gastric Immune and Inflammatory Disorders: The Evidence and Controversies. Diseases 2015, 3, 34–55. [Google Scholar] [CrossRef] [PubMed]
- Pellicano, R.; Ianiro, G.; Fagoonee, S.; Settanni, C.R.; Gasbarrini, A. Review: Extragastric Diseases and Helicobacter pylori. Helicobacter 2020, 25, e12741. [Google Scholar]
- Yu, Y.; Zhu, S.; Li, P.; Min, L.; Zhang, S. Helicobacter pylori Infection and Inflammatory Bowel Disease: A Crosstalk between Upper and Lower Digestive Tract. Cell Death Dis. 2018, 9, 961. [Google Scholar] [CrossRef]
- Ansari, S.; Yamaoka, Y. Helicobacter pylori Virulence Factors Exploiting Gastric Colonization and Its Pathogenicity. Toxins 2019, 11, 677. [Google Scholar] [CrossRef] [Green Version]
- Percival, S.L.; Suleman, L. Biofilms and Helicobacter pylori: Dissemination and Persistence within the Environment and Host. World J. Gastrointest. Pathophysiol. 2014, 5, 122–132. [Google Scholar]
- Stark, R.M.; Gerwig, G.J.; Pitman, R.S.; Potts, L.F.; Williams, N.A.; Greenman, J.; Weinzweig, I.P.; Hirst, T.R.; Millar, M.R. Biofilm Formation by Helicobacter pylori. Lett. Appl. Microbiol. 1999, 28, 121–126. [Google Scholar] [CrossRef]
- Hathroubi, S.; Zerebinski, J.; Ottemann, K.M. Helicobacter pylori Biofilm Involves a Multigene Stress-Biased Response, Including a Structural Role for Flagella. mBio 2018, 9, e01973-18. [Google Scholar] [CrossRef] [Green Version]
- Hathroubi, S.; Zerebinski, J.; Ottemann, K.M. Helicobacter pylori Biofilm Cells are Metabolically Distinct, Express Flagella, and Antibiotic Tolerant. bioRxiv 2019. [Google Scholar] [CrossRef]
- Fauzia, K.A.; Miftahussurur, M.; Syam, A.F.; Waskito, L.A.; Doohan, D.; Rezkitha, Y.A.A.; Matsumoto, T.; Tuan, V.P.; Akada, J.; Yonezawa, H.; et al. Biofilm Formation and Antibiotic Resistance Phenotype of Helicobacter pylori Clinical Isolates. Toxins 2020, 12, 473. [Google Scholar] [CrossRef]
- Windham, I.H.; Servetas, S.L.; Whitmire, J.M.; Pletzer, D.; Hancock, R.E.W.; Merrell, D.S. Helicobacter pylori Biofilm Formation Is Differentially Affected by Common Culture Conditions, and Proteins Play a Central Role in the Biofilm Matrix. Appl. Environ. Microbiol. 2018, 84, e00391-18. [Google Scholar]
- Grande, R.; Di Marcantonio, M.C.; Robuffo, I.; Pompilio, A.; Celia, C.; Di Marzio, L.; Paolino, D.; Codagnone, M.; Muraro, R.; Stoodley, P.; et al. Helicobacter pylori ATCC 43629/NCTC 11639 Outer Membrane Vesicles (OMVs) from Biofilm and Planktonic Phase Associated with Extracellular DNA (eDNA). Front. Microbiol. 2015, 6, 1369. [Google Scholar] [CrossRef] [Green Version]
- Anderson, J.K.; Huang, J.Y.; Wreden, C.; Sweeney, E.G.; Goers, J.; Remington, S.J.; Guillemin, K. Chemorepulsion from the Quorum Signal Autoinducer-2 Promotes Helicobacter pylori Biofilm Dispersal. mBio 2015, 6, e00379. [Google Scholar] [CrossRef] [Green Version]
- Tan, S.; Tompkins, L.S.; Amieva, M.R. Helicobacter pylori Usurps Cell Polarity to Turn the Cell Surface into a Replicative Niche. PLoS Pathog. 2009, 5, e1000407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Attaran, B.; Falsafi, T.; Moghaddam, A.N. Study of Biofilm Formation in C57Bl/6J Mice by Clinical Isolates of Helicobacter pylori. Saudi J. Gastroenterol. 2016, 22, 161–168. [Google Scholar]
- Carron, M.A.; Tran, V.R.; Sugawa, C.; Coticchia, J.M. Identification of Helicobacter pylori Biofilms in Human Gastric Mucosa. J. Gastrointest. Surg. 2006, 10, 712–717. [Google Scholar] [CrossRef]
- Coticchia, J.M.; Sugawa, C.; Tran, V.R.; Gurrola, J.; Kowalski, E.; Carron, M.A. Presence and Density of Helicobacter pylori Biofilms in Human Gastric Mucosa in Patients With Peptic Ulcer Disease. J. Gastrointest. Surg. 2006, 10, 883–889. [Google Scholar] [CrossRef]
- Ng, C.G.; Loke, M.F.; Goh, K.L.; Vadivelu, J.; Ho, B. Biofilm Formation Enhances Helicobacter pylori Survivability in Vegetables. Food Microbiol. 2017, 62, 68–76. [Google Scholar] [CrossRef]
- Linke, S.; Lenz, J.; Gemein, S.; Exner, M.; Gebel, J. Detection of Helicobacter pylori in Biofilms by Real-Time PCR. Int. J. Hyg. Environ. Health 2010, 213, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Park, S.R.; Mackay, W.G.; Reid, D.C. Helicobacter sp. Recovered from Drinking Water Biofilm Sampled from a Water Distribution System. Water Res. 2001, 35, 1624–1626. [Google Scholar] [CrossRef]
- Bunn, J.E.G.; MacKay, W.G.; Thomas, J.E.; Reid, D.C.; Weaver, L.T. Detection of Helicobacter pylori DNA in Drinking Water Biofilms: Implications for Transmission in Early Life. Lett. Appl. Microbiol. 2002, 34, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Percival, S.L.; Thomas, J.G. Transmission of Helicobacter pylori and the Role of Water and Biofilms. J. Water Health 2009, 7, 469–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hathroubi, S.; Zerebinski, J.; Clarke, A.; Ottemann, K.M. Helicobacter pylori Biofilm Confers Antibiotic Tolerance in Part via a Protein-Dependent Mechanism. Antibiotics 2020, 9, 355. [Google Scholar] [CrossRef]
- Yonezawa, H.; Osaki, T.; Hojo, F.; Kamiya, S. Effect of Helicobacter pylori Biofilm Formation on Susceptibility to Amoxicillin, Metronidazole and Clarithromycin. Microb. Pathog. 2019, 132, 100–108. [Google Scholar] [CrossRef]
- Cai, Y.; Wang, C.; Chen, Z.; Xu, Z.; Li, H.; Li, W.; Sun, Y. Transporters HP0939, HP0497, and HP0471 Participate in Intrinsic Multidrug Resistance and Biofilm Formation in Helicobacter pylori by Enhancing Drug Efflux. Helicobacter 2020, 25, e12715. [Google Scholar] [CrossRef]
- Ge, X.; Cai, Y.; Chen, Z.; Gao, S.; Geng, X.; Li, Y.; Li, Y.; Jia, J.; Sun, Y. Bifunctional Enzyme SpoT Is Involved in Biofilm Formation of Helicobacter pylori with Multidrug Resistance by Upregulating Efflux Pump Hp1174 (gluP). Antimicrob. Agents Chemother. 2018, 62, e00957-18. [Google Scholar] [CrossRef] [Green Version]
- Cole, S.P.; Harwood, J.; Lee, R.; She, R.; Guiney, D.G. Characterization of Monospecies Biofilm Formation by Helicobacter pylori. J. Bacteriol. 2004, 186, 3124–3132. [Google Scholar] [CrossRef] [Green Version]
- Servetas, S.L.; Carpenter, B.M.; Haley, K.P.; Gilbreath, J.J.; Gaddy, J.A.; Scott Merrell, D. Characterization of Key Helicobacter pylori Regulators Identifies a Role for ArsRS in Biofilm Formation. J. Bacteriol. 2016, 198, 2536–2548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ratthawongjirakul, P.; Thongkerd, V.; Chaicumpa, W. The Impacts of a fliD Mutation on the Biofilm Formation of Helicobacter pylori. Asian Pac. J. Trop. Biomed. 2016, 6, 1008–1014. [Google Scholar] [CrossRef] [Green Version]
- Shao, C.; Sun, Y.; Wang, N.; Yu, H.; Zhou, Y.; Chen, C.; Jia, J. Changes of Proteome Components of Helicobacter pylori Biofilms Induced by Serum Starvation. Mol. Med. Rep. 2013, 8, 1761–1766. [Google Scholar] [CrossRef] [Green Version]
- Krzyżek, P.; Gościniak, G.; Fijałkowski, K.; Migdał, P.; Dziadas, M.; Owczarek, A.; Czajkowska, J.; Aniołek, O.; Junka, A. Potential of Bacterial Cellulose Chemisorbed with Anti-Metabolites, 3-Bromopyruvate or Sertraline, to Fight against Helicobacter pylori Lawn Biofilm. Int. J. Mol. Sci. 2020, 21, 9507. [Google Scholar] [CrossRef]
- Azevedo, N.F.; Pinto, A.R.; Reis, N.M.; Vieira, M.J.; Keevil, C.W. Shear Stress, Temperature, and Inoculation Concentration Influence the Adhesion of Water-Stressed Helicobacter pylori to Stainless Steel 304 and Polypropylene. Appl. Environ. Microbiol. 2006, 72, 2936–2941. [Google Scholar] [CrossRef] [Green Version]
- Bessa, L.J.; Grande, R.; DI Iorio, D.; DI Giulio, M.; DI Campli, E.; Cellini, L. Helicobacter pylori Free-Living and Biofilm Modes of Growth: Behavior in Response to Different Culture Media. APMIS 2013, 121, 549–560. [Google Scholar] [CrossRef]
- Cárdenas-Mondragón, M.G.; Ares, M.A.; Panunzi, L.G.; Pacheco, S.; Camorlinga-Ponce, M.; Girón, J.A.; Torres, J.; De la Cruz, M.A. Transcriptional Profiling of Type II Toxin-Antitoxin Genes of Helicobacter pylori under Different Environmental Conditions: Identification of HP0967-HP0968 System. Front. Microbiol. 2016, 7, 1872. [Google Scholar] [CrossRef] [Green Version]
- Sweeney, E.G.; Nishida, A.; Weston, A.; Bañuelos, M.S.; Potter, K.; Conery, J.; Guillemin, K. Agent-Based Modeling Demonstrates How Local Chemotactic Behavior Can Shape Biofilm Architecture. mSphere 2019, 4, e00285-19. [Google Scholar] [CrossRef] [Green Version]
- Hung, C.L.; Cheng, H.H.; Hsieh, W.C.; Tsai, Z.T.Y.; Tsai, H.K.; Chu, C.H.; Hsieh, W.P.; Chen, Y.F.; Tsou, Y.; Lai, C.H.; et al. The CrdRS Two-Component System in Helicobacter pylori Responds to Nitrosative Stress. Mol. Microbiol. 2015, 97, 1128–1141. [Google Scholar] [CrossRef]
- De la Cruz, M.A.; Ares, M.A.; von Bargen, K.; Panunzi, L.G.; Martínez-Cruz, J.; Valdez-Salazar, H.A.; Jiménez-Galicia, C.; Torres, J. Gene Expression Profiling of Transcription Factors of Helicobacter pylori under Different Environmental Conditions. Front. Microbiol. 2017, 8, 615. [Google Scholar] [CrossRef] [Green Version]
- Wong, E.H.J.; Ng, C.G.; Chua, E.G.; Tay, A.C.Y.; Peters, F.; Marshall, B.J.; Ho, B.; Goh, K.L.; Vadivelu, J.; Loke, M.F. Comparative Genomics Revealed Multiple Helicobacter pylori Genes Associated with Biofilm Formation In Vitro. PLoS ONE 2016, 11, e0166835. [Google Scholar] [CrossRef] [PubMed]
- Servetas, S.L.; Doster, R.S.; Kim, A.; Windham, I.H.; Cha, J.H.; Gaddy, J.A.; Merrell, D.S. ArsRS-Dependent Regulation of homB Contributes to Helicobacter pylori Biofilm Formation. Front. Microbiol. 2018, 9, 1497. [Google Scholar] [CrossRef] [PubMed]
- Yonezawa, H.; Osaki, T.; Hanawa, T.; Kurata, S.; Ochiai, K.; Kamiya, S. Impact of Helicobacter pylori Biofilm Formation on Clarithromycin Susceptibility and Generation of Resistance Mutations. PLoS ONE 2013, 8, e73301. [Google Scholar] [CrossRef] [PubMed]
- Attaran, B.; Falsafi, T.; Ghorbanmehr, N. Effect of Biofilm Formation by Clinical Isolates of Helicobacter pylori on the Efflux-Mediated Resistance to Commonly Used Antibiotics. World J. Gastroenterol. 2017, 23, 1163–1170. [Google Scholar] [CrossRef] [PubMed]
- Kazakos, E.I.; Dorrell, N.; Polyzos, S.A.; Deretzi, G.; Kountouras, J. Comment on “Effect of Biofilm Formation by Clinical Isolates of Helicobacter pylori on the Efflux-Mediated Resistance to Commonly Used Antibiotics. ” World J. Gastroenterol. 2017, 23, 6194–6196. [Google Scholar] [CrossRef]
- Yang, F.L.; Hassanbhai, A.M.; Chen, H.Y.; Huang, Z.Y.; Lin, T.L.; Wu, S.H.; Ho, B. Proteomannans in Biofilm of Helicobacter pylori ATCC 43504. Helicobacter 2011, 16, 89–98. [Google Scholar] [CrossRef]
- Wong, E.H.J.; Ng, C.G.; Goh, K.L.; Vadivelu, J.; Ho, B.; Loke, M.F. Metabolomic Analysis of Low and High Biofilm-Forming Helicobacter pylori Strains. Sci. Rep. 2018, 8, 1409. [Google Scholar] [CrossRef] [Green Version]
- Cellini, L.; Grande, R.; Traini, T.; Di Campli, E.; Di Bartolomeo, S.; Di Iorio, D.; Caputi, S. Biofilm Formation and Modulation of luxS and rpoD Expression by Helicobacter pylori. Biofilms 2005, 2, 119–127. [Google Scholar] [CrossRef]
- Cellini, L.; Grande, R.; Di Campli, E.; Di Bartolomeo, S.; Di Giulio, M.; Traini, T.; Trubiani, O. Characterization of an Helicobacter pylori Environmental Strain. J. Appl. Microbiol. 2008, 105, 761–769. [Google Scholar] [CrossRef]
- Bervoets, I.; Charlier, D. Diversity, Versatility and Complexity of Bacterial Gene Regulation Mechanisms: Opportunities and Drawbacks for Applications in Synthetic Biology. FEMS Microbiol. Rev. 2019, 43, 304–339. [Google Scholar] [CrossRef] [Green Version]
- Balleza, E.; López-Bojorquez, L.N.; Martínez-Antonio, A.; Resendis-Antonio, O.; Lozada-Chávez, I.; Balderas-Martínez, Y.I.; Encarnación, S.; Collado-Vides, J. Regulation by Transcription Factors in Bacteria: Beyond Description. FEMS Microbiol. Rev. 2009, 33, 133–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Austin, C.M.; Wang, G.; Maier, R.J. Aconitase Functions as a Pleiotropic Posttranscriptional Regulator in Helicobacter pylori. J. Bacteriol. 2015, 197, 3076–3086. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Romero-Gallo, J.; Benoit, S.L.; Blanca Piazuelo, M.; Dominguez, R.L.; Morgan, D.R.; Peek, R.M.; Maier, R.J. Hydrogen Metabolism in Helicobacter pylori Plays a Role in Gastric Carcinogenesis through Facilitating CagA Translocation. mBio 2016, 7, e01022-16. [Google Scholar] [CrossRef] [Green Version]
- Kuhns, L.G.; Benoit, S.L.; Bayyareddy, K.; Johnson, D.; Orlando, R.; Evans, A.L.; Waldrop, G.L.; Maier, R.J. Carbon Fixation Driven by Molecular Hydrogen Results in Chemolithoautotrophically Enhanced Growth of Helicobacter pylori. J. Bacteriol. 2016, 198, 1423–1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krüger, N.J.; Knüver, M.T.; Zawilak-Pawlik, A.; Appel, B.; Stingl, K. Genetic Diversity as Consequence of a Microaerobic and Neutrophilic Lifestyle. PLoS Pathog. 2016, 12, e1005626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roncarati, D.; Danielli, A.; Spohn, G.; Delany, I.; Scarlato, V. Transcriptional Regulation of Stress Response and Motility Functions in Helicobacter pylori is Mediated by HspR and HrcA. J. Bacteriol. 2007, 189, 7234–7243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roncarati, D.; Pinatel, E.; Fiore, E.; Peano, C.; Loibman, S.; Scarlato, V. Helicobacter pylori Stress-Response: Definition of the HrcA Regulon. Microorganisms 2019, 7, 436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Häuser, R.; Pech, M.; Kijek, J.; Yamamoto, H.; Titz, B.; Naeve, F.; Tovchigrechko, A.; Yamamoto, K.; Szaflarski, W.; Takeuchi, N.; et al. RsFA (YbeB) Proteins are Conserved Ribosomal Silencing Factors. PLoS Genet. 2012, 8, e1002815. [Google Scholar] [CrossRef] [Green Version]
- Acio-Pizzarello, C.R.; Acio, A.A.; Choi, E.J.; Bond, K.; Kim, J.; Kenan, A.C.; Chen, J.; Forsyth, M.H. Determinants of the Regulation of Helicobacter pylori Adhesins Include Repeat Sequences in both Promoter and Coding Regions as Well as the Two-Component System ArsRS. J. Med. Microbiol. 2017, 66, 798–807. [Google Scholar] [CrossRef]
- Krzyżek, P.; Gościniak, G. Morphology of Helicobacter pylori as a Result of Peptidoglycan and Cytoskeleton Rearrangements. Prz. Gastroenterol. 2018, 13, 182–195. [Google Scholar] [CrossRef] [Green Version]
- Krzyżek, P.; Grande, R. Transformation of Helicobacter pylori into Coccoid Forms as a Challenge for Research Determining Activity of Antimicrobial Substances. Pathogens 2020, 9, 184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krzyżek, P.; Biernat, M.M.; Gościniak, G. Intensive Formation of Coccoid Forms as a Feature Strongly Associated with Highly Pathogenic Helicobacter pylori Strains. Folia Microbiol. 2019, 64, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Dong, K.; Pan, H.; Yang, D.; Rao, L.; Zhao, L.; Wang, Y.; Liao, X. Induction, Detection, Formation, and Resuscitation of Viable but Non-Culturable State Microorganisms. Compr. Rev. Food Sci. Food Saf. 2020, 19, 149–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kadkhodaei, S.; Siavoshi, F.; Akbari Noghabi, K. Mucoid and Coccoid Helicobacter pylori with Fast Growth and Antibiotic Resistance. Helicobacter 2019, 25, e12678. [Google Scholar] [CrossRef]
- Poursina, F.; Fagri, J.; Mirzaei, N.; Safaei, H.G. Overexpression of spoT Gene in Coccoid Forms of Clinical Helicobacter pylori Isolates. Folia Microbiol. 2018, 63, 459–465. [Google Scholar] [CrossRef]
- Geng, X.; Li, W.; Chen, Z.; Gao, S.; Hong, W.; Ge, X.; Hou, G.; Hu, Z.; Zhou, Y.; Zeng, B.; et al. The Bifunctional Enzyme SpoT Is Involved in the Clarithromycin Tolerance of Helicobacter pylori by Upregulating the Transporters HP0939, HP1017, HP0497, and HP0471. Antimicrob. Agents Chemother. 2017, 61, e02011-16. [Google Scholar] [CrossRef] [Green Version]
- Wuchty, S.; Müller, S.A.; Caufield, J.H.; Häuser, R.; Aloy, P.; Kalkhof, S.; Uetz, P. Proteome Data Improves Protein Function Prediction in the Interactome of Helicobacter pylori. Mol. Cell. Proteom. 2018, 17, 961–973. [Google Scholar] [CrossRef] [Green Version]
- Loke, M.F.; Ng, C.G.; Vilashni, Y.; Lim, J.; Ho, B. Understanding the Dimorphic Lifestyles of Human Gastric Pathogen Helicobacter pylori Using the SWATH-Based Proteomics Approach. Sci. Rep. 2016, 6, 26784. [Google Scholar] [CrossRef]
- Benoit, S.L.; Seshadri, S.; Lamichhane-Khadka, R.; Maier, R.J. Helicobacter hepaticus NikR Controls Urease and Hydrogenase Activities via the NikABDE and HH0418 Putative Nickel Import Proteins. Microbiology 2013, 159, 136–146. [Google Scholar] [CrossRef] [Green Version]
- Shariq, M.; Kumar, N.; Kumari, R.; Kumar, A.; Subbarao, N.; Mukhopadhyay, G. Biochemical Analysis of CagE: A VirB4 Homologue of Helicobacter pylori Cag-T4SS. PLoS ONE 2015, 10, e0142606. [Google Scholar] [CrossRef]
- Terebiznik, M.R.; Vazquez, C.L.; Torbicki, K.; Banks, D.; Wang, T.; Hong, W.; Blanke, S.R.; Colombo, M.I.; Jones, N.L. Helicobacter pylori VacA toxin promotes bacterial intracellular survival in gastric epithelial cells. Infect. Immun. 2006, 74, 6599–6614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vallese, F.; Mishra, N.M.; Pagliari, M.; Berto, P.; Codolo, G.; de Bernard, M.; Zanotti, G. Helicobacter pylori Antigenic Lpp20 is a Structural Homologue of Tipα and Promotes Epithelial-Mesenchymal Transition. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 3263–3271. [Google Scholar] [CrossRef] [PubMed]
- Zepeda Gurrola, R.C.; Fu, Y.; Rodríguez Luna, I.C.; Benítez Cardoza, C.G.; de Jesús López López, M.; López Vidal, Y.; Gutíerrez, G.R.A.; Rodríguez Pérez, M.A.; Guo, X. Novel protein interactions with an actin homolog (MreB) of Helicobacter pylori determined by bacterial two-hybrid system. Microbiol. Res. 2017, 201, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Guo, G.; Mao, X.H.; De Tong, W.; Zou, Q.M. Proteomic Insights into Helicobacter pylori Coccoid Forms under Oxidative Stress. Curr. Microbiol. 2008, 57, 281–286. [Google Scholar] [CrossRef]
- Attaran, B.; Falsafi, T. Identification of Factors Associated with Biofilm Formation Ability in the Clinical Isolates of Helicobacter pylori. Iran. J. Biotechnol. 2017, 15, 58–66. [Google Scholar] [CrossRef] [Green Version]
- Smith, W.P.J.; Davit, Y.; Osborne, J.M.; Kim, W.D.; Foster, K.R.; Pitt-Francis, J.M. Cell Morphology Drives Spatial Patterning in Microbial Communities. Proc. Natl. Acad. Sci. USA 2017, 114, E280–E286. [Google Scholar] [CrossRef] [Green Version]
- Serra, D.O.; Richter, A.M.; Klauck, G.; Mika, F.; Hengge, R. Microanatomy at Cellular Resolution and Spatial Order of Physiological Differentiation in a Bacterial Biofilm. mBio 2013, 4, 103–116. [Google Scholar] [CrossRef] [Green Version]
- Bastian, F.O.; Elzer, P.H.; Wu, X. Spiroplasma spp. Biofilm Formation Is Instrumental for Their Role in the Pathogenesis of Plant, Insect and Animal Diseases. Exp. Mol. Pathol. 2012, 93, 116–128. [Google Scholar] [CrossRef]
- Peschek, N.; Herzog, R.; Singh, P.K.; Sprenger, M.; Meyer, F.; Fröhlich, K.S.; Schröger, L.; Bramkamp, M.; Drescher, K.; Papenfort, K. RNA-Mediated Control of Cell Shape Modulates Antibiotic Resistance in Vibrio cholerae. Nat. Commun. 2020, 11, 6067. [Google Scholar] [CrossRef]
- Ferreira, J.A.; Pires, C.; Paulo, M.; Azevedo, N.F.; Domingues, M.R.; Vieira, M.J.; Monteiro, M.A.; Coimbra, M.A. Bioaccumulation of Amylose-Like Glycans by Helicobacter pylori. Helicobacter 2009, 14, 559–570. [Google Scholar] [CrossRef] [Green Version]
- Fu, H.W. Helicobacter pylori Neutrophil-Activating Protein: From Molecular Pathogenesis to Clinical Applications. World J. Gastroenterol. 2014, 20, 5294–5301. [Google Scholar] [CrossRef] [PubMed]
- Schwechheimer, C.; Kuehn, M.J. Outer-Membrane Vesicles from Gram-Negative Bacteria: Biogenesis and Functions. Nat. Rev. Microbiol. 2015, 13, 619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jan, A.T. Outer Membrane Vesicles (OMVs) of Gram-negative Bacteria: A Perspective Update. Front. Microbiol. 2017, 8, 1053. [Google Scholar] [CrossRef] [PubMed]
- Nagakubo, T.; Nomura, N.; Toyofuku, M. Cracking Open Bacterial Membrane Vesicles. Front. Microbiol. 2020, 10, 3026. [Google Scholar] [CrossRef] [Green Version]
- Krzyżek, P. Sekrecja Pęcherzyków Błonowych Jako Mechanizm Promujący Infekcje H. pylori. Postępy Mikrobiol. 2017, 56, 316–325. [Google Scholar] [CrossRef]
- Jarzab, M.; Posselt, G.; Meisner-Kober, N.; Wessler, S. Helicobacter pylori-Derived Outer Membrane Vesicles (OMVs): Role in Bacterial Pathogenesis? Microorganisms 2020, 8, 1328. [Google Scholar] [CrossRef]
- Grande, R.; Di Giulio, M.; Bessa, L.J.; Di Campli, E.; Baffoni, M.; Guarnieri, S.; Cellini, L. Extracellular DNA in Helicobacter pylori Biofilm: A Backstairs Rumour. J. Appl. Microbiol. 2011, 110, 490–498. [Google Scholar]
- Puca, V.; Ercolino, E.; Celia, C.; Bologna, G.; Di Marzio, L.; Mincione, G.; Marchisio, M.; Miscia, S.; Muraro, R.; Lanuti, P.; et al. Detection and Quantification of eDNA-Associated Bacterial Membrane Vesicles by Flow Cytometry. Int. J. Mol. Sci. 2019, 20, 5307. [Google Scholar] [CrossRef] [Green Version]
- Yonezawa, H.; Osaki, T.; Kurata, S.; Fukuda, M.; Kawakami, H.; Ochiai, K.; Hanawa, T.; Kamiya, S. Outer Membrane Vesicles of Helicobacter pylori TK1402 are Involved in Biofilm Formation. BMC Microbiol. 2009, 9, 197. [Google Scholar] [CrossRef] [Green Version]
- Yonezawa, H.; Osaki, T.; Woo, T.; Kurata, S.; Zaman, C.; Hojo, F.; Hanawa, T.; Kato, S.; Kamiya, S. Analysis of Outer Membrane Vesicle Protein Involved in Biofilm Formation of Helicobacter pylori. Anaerobe 2011, 17, 390. [Google Scholar] [CrossRef]
- Yonezawa, H.; Osaki, T.; Fukutomi, T.; Hanawa, T.; Kurata, S.; Zaman, C.; Hojo, F.; Kamiya, S. Diversification of the AlpB Outer Membrane Protein of Helicobacter pylori Affects Biofilm Formation and Cellular Adhesion. J. Bacteriol. 2017, 199, e00729-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zavan, L.; Bitto, N.J.; Johnston, E.L.; Greening, D.W.; Kaparakis-Liaskos, M. Helicobacter pylori Growth Stage Determines the Size, Protein Composition, and Preferential Cargo Packaging of Outer Membrane Vesicles. Proteomics 2019, 19, e1800209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, L.; Bitto, N.J.; Steer, D.L.; Lo, C.; D’Costa, K.; Ramm, G.; Shambrook, M.; Hill, A.F.; Ferrero, R.L.; Kaparakis-Liaskos, M. Helicobacter pylori Outer Membrane Vesicle Size Determines their Mechanisms of Host Cell Entry and Protein Content. Front. Immunol. 2018, 9, 1466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lekmeechai, S.; Su, Y.-C.; Brant, M.; Alvarado-Kristensson, M.; Vallström, A.; Obi, I.; Arnqvist, A.; Riesbeck, K. Helicobacter pylori Outer Membrane Vesicles Protect the Pathogen from Reactive Oxygen Species of the Respiratory Burst. Front. Microbiol. 2018, 9, 1837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, B.O.; Dawson, R.A.; Alsharaf, L.M.; Winter, J.A. Protective Effects of Helicobacter pylori Membrane Vesicles against Stress and Antimicrobial Agents. Microbiology 2020, 166, 751–758. [Google Scholar] [CrossRef]
- Blanco, P.; Hernando-Amado, S.; Reales-Calderon, J.; Corona, F.; Lira, F.; Alcalde-Rico, M.; Bernardini, A.; Sanchez, M.; Martinez, J. Bacterial multidrug efflux pumps: Much more than antibiotic resistance determinants. Microorganisms 2016, 4, 14. [Google Scholar] [CrossRef] [Green Version]
- Lekshmi, M.; Parvathi, A.; Kumar, S.; Varela, M.F. Efflux pump-mediated quorum sensing: New avenues for modulation of antimicrobial resistance and bacterial virulence. In Biotechnological Applications of Quorum Sensing Inhibitors; Springer: Singapore, 2018; pp. 127–142. ISBN 9789811090264. [Google Scholar]
- Alav, I.; Sutton, J.M.; Rahman, K.M. Role of Bacterial Efflux Pumps in Biofilm Formation. J. Antimicrob. Chemother. 2018, 73, 2003–2020. [Google Scholar] [CrossRef] [Green Version]
- Navarro Llorens, J.M.; Tormo, A.; Martínez-García, E. Stationary Phase in Gram-Negative Bacteria. FEMS Microbiol. Rev. 2010, 34, 476–495. [Google Scholar] [CrossRef] [Green Version]
- Federle, M.J. Autoinducer-2-Based Chemical Communication in Bacteria: Complexities of Interspecies Signaling. Contrib. Microbiol. 2009, 16, 18–32. [Google Scholar]
- Pereira, C.S.; Thompson, J.A.; Xavier, K.B. AI-2-Mediated Signalling in Bacteria. FEMS Microbiol. Rev. 2013, 37, 156–181. [Google Scholar] [CrossRef] [Green Version]
- Shen, F.; Hobley, L.; Doherty, N.; Loh, J.T.; Cover, T.L.; Sockett, R.E.; Hardie, K.R.; Atherton, J.C. In Helicobacter pylori Auto-inducer-2, but not LuxS/MccAB Catalysed Reverse Transsulphuration, Regulates Motility through Modulation of Flagellar Gene Transcription. BMC Microbiol. 2010, 10, 210. [Google Scholar] [CrossRef] [PubMed]
- Xia, X. Protein isoelectric point and Helicobacter pylori. In Bioinformatics and the Cell; Springer International Publishing: Berlin/Heidelberg, Germany, 2018; pp. 397–412. [Google Scholar]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krzyżek, P.; Grande, R.; Migdał, P.; Paluch, E.; Gościniak, G. Biofilm Formation as a Complex Result of Virulence and Adaptive Responses of Helicobacter pylori. Pathogens 2020, 9, 1062. https://doi.org/10.3390/pathogens9121062
Krzyżek P, Grande R, Migdał P, Paluch E, Gościniak G. Biofilm Formation as a Complex Result of Virulence and Adaptive Responses of Helicobacter pylori. Pathogens. 2020; 9(12):1062. https://doi.org/10.3390/pathogens9121062
Chicago/Turabian StyleKrzyżek, Paweł, Rossella Grande, Paweł Migdał, Emil Paluch, and Grażyna Gościniak. 2020. "Biofilm Formation as a Complex Result of Virulence and Adaptive Responses of Helicobacter pylori" Pathogens 9, no. 12: 1062. https://doi.org/10.3390/pathogens9121062
APA StyleKrzyżek, P., Grande, R., Migdał, P., Paluch, E., & Gościniak, G. (2020). Biofilm Formation as a Complex Result of Virulence and Adaptive Responses of Helicobacter pylori. Pathogens, 9(12), 1062. https://doi.org/10.3390/pathogens9121062