Characterization of the Proteolytic Activity of a Halophilic Aspergillus reticulatus Strain SK1-1 Isolated from a Solar Saltern
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection, Isolation, and Cultivation of Fungi
2.2. Optimization of NaCl Concentrations for Radial Growth of SK1-1
2.3. Identification of SK1-1
2.4. Phylogenetic Analysis
2.5. Skim Milk Agar Assay for Proteolytic Activity
2.6. Preparation of the Extracellular Crude Enzyme
2.7. Proteolytic Activity Assays Using Azocasein as a Substrate
2.8. Influence of Temperature, pH, and NaCl Concentration on Proteolytic Activity
2.9. Influence of Metal Ions on Proteolytic Activity
2.10. Determination of Thermal Stability of Proteolytic Activity
2.11. Protease Inhibitor Assay
2.12. Statistical Analysis
3. Results
3.1. Halophilic Growth of SK1-1
3.2. Identification of SK1-1
3.3. Skim Milk Agar Assay of SK1-1
3.4. Construction of Calibration Curve for Azocasein Assays
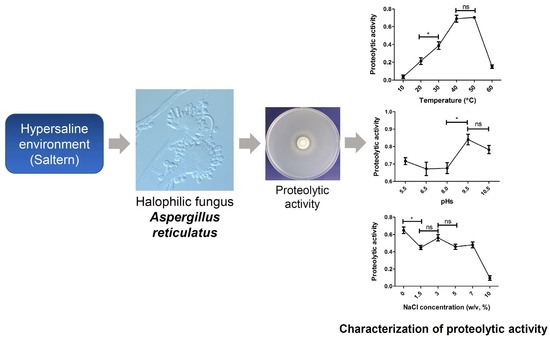
3.5. Influence of Temperature, pH, and NaCl Concentration on Proteolytic Activity of SK1-1
3.6. Effects of Metal Ions on Proteolytic Activity of SK1-1
3.7. Thermal Stability of the Crude Enzymes of SK1-1
3.8. Protease Inhibitor Assay
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gostincar, C.; Grube, M.; de Hoog, S.; Zalar, P.; Gunde-Cimerman, N. Extremotolerance in fungi: Evolution on the edge. FEMS Microbiol. Ecol. 2010, 71, 2–11. [Google Scholar] [CrossRef] [Green Version]
- Dumome, K.; Cordova, D.C.; Astorga-Elo, M.; Renganathan, P. Extremozymes: A potential source for industrial applications. J. Microbiol. Biotechnol. 2017, 27, 649–659. [Google Scholar]
- Marrs, B.; Delagrave, S.; Murphy, D. Novel approaches for discovering industrial enzymes. Curr. Opin. Microbiol. 1999, 2, 241–245. [Google Scholar] [CrossRef]
- Chung, D.; Kim, H.; Choi, H.S. Fungi in salterns. J. Microbiol. 2019, 57, 717–724. [Google Scholar] [CrossRef] [PubMed]
- Gunde-Cimerman, N.; Zalar, P.; Hoog, S.; Plemenitas, A. Hypersaline waters in salterns—Natural ecological niches for halophilic black yeasts. FEMS Microbiol. Ecol. 2000, 32, 235–240. [Google Scholar]
- Cantrell, S.A.; Casillas-Martinez, L.; Molina, M. Characterization of fungi from hypersaline environments of solar salterns using morphological and molecular techniques. Mycol. Res. 2006, 110, 962–970. [Google Scholar] [CrossRef] [PubMed]
- Butinar, L.; Frisvad, J.C.; Gunde-Cimerman, N. Hypersaline waters—A potential source of foodborne toxigenic aspergilli and penicillia. FEMS Microbiol. Ecol. 2011, 77, 186–199. [Google Scholar] [CrossRef]
- Zalar, P.; De Hoog, G.S.; Schroers, H.J.; Frank, J.M.; Gunde-Cimerman, N. Taxonomy and phylogeny of the xerophilic genus Wallemia (Wallemiomycetes and Wallemiales, cl. et ord. nov.). Antonie Leeuwenhoek 2005, 87, 311–328. [Google Scholar] [CrossRef]
- Nazareth, S.; Gonsalves, V. Aspergillus penicillioides—A true halophile existing in hypersaline and polyhaline econiches. Ann. Microbiol. 2013, 64, 397–402. [Google Scholar] [CrossRef]
- Shanmugavel, M.; Vasantharaj, S.; Saathiyavimal, S.; Gnanamani, A. Application of an alkaline protease in biological waste processing: An eco-friendly approach. Int. J. Biosci. Nanosci. 2016, 3, 19–24. [Google Scholar]
- de Souza, P.M.; de Assis Bittencourt, M.L.; Caprara, C.C.; de Freitas, M.; de Almeida, R.P.C.; Silveira, D.; Fonseca, Y.M.; Filho, E.X.F.; Pessoa, A., Jr.; Magalhaes, P.O. A biotechnology perspective of fungal proteases. Braz. J. Microbiol. 2015, 46, 337–346. [Google Scholar] [CrossRef] [Green Version]
- Kwon, Y.M.; Choi, H.S.; Lim, J.Y.; Jang, H.S.; Chung, D. Characterization of amylolytic activity by a marine-derived yeast Sporidiobolus pararoseus PH-Gra1. Mycobiology 2020, 48, 195–203. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Glass, N.L.; Donaldson, G.C. Development of premier sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [Green Version]
- Coelho, D.F.; Saturnino, T.P.; Fernandes, F.F.; Mazzola, P.G.; Silveira, E.; Tambourgi, E.B. Azocasein substrate for determination of proteolytic activity: Reexamining a traditional method using bromelain samples. BioMed Res. Int. 2016, 2016, 8409183. [Google Scholar] [CrossRef] [Green Version]
- Han, C.; Kautto, L.; Nevalainen, H. Secretion of proteases by an opportunistic fungal pathogen Scedosporium aurantiacum. PLoS ONE 2017, 12, e0169403. [Google Scholar]
- Kushiner, D.J. Life in high salt and solute concentrations. In Microbial Life in Extreme Environments; Kushiner, D.J., Ed.; Academic Press: London, UK, 1978; pp. 317–368. [Google Scholar]
- Larsen, H. Halophilic and halotolerant microorganism—An overview and historical perspective. FEMS Microbiol. Rev. 1986, 2, 3–7. [Google Scholar] [CrossRef]
- Sklenar, F.; Jurjevic, Z.; Zalar, P.; Frisvad, J.C.; Visagie, C.M.; Kolarik, M.; Houbraken, J.; Chen, A.J.; Yilmaz, N.; Seifert, K.A.; et al. Phylogeny of xerophilic aspergilli (subgenus Aspergillus) and taxonomic revision of section Restricti. Stud. Mycol. 2017, 88, 161–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, I.; Siwarungson, N.; Punnapayak, H.; Lotrakul, P.; Prasongsuk, S.; Bankeeree, W.; Rakshit, S.K. Screening of potential biotechnological applications from obligate halophilic fungi, isolated from a man-made solar saltern located in Phetchaburi province, Thailand. Pak. J. Bot. 2014, 46, 983–988. [Google Scholar]
- Chamekh, R.; Deniel, F.; Donot, C.; Jany, J.-L.; Nodet, P.; Belabid, L. Isolation, identification and enzymatic activity of halotolerant and halophilic fungi from the Great Sebkha of Oran in northwestern of Algeria. Mycobiology 2019, 47, 230–241. [Google Scholar] [CrossRef]
- Ali, I.; Akbar, A.; Yanwisetpakdee, B.; Prasongsuk, S.; Lotrakul, P.; Punnapayak, H. Purification, characterization, and potential of saline waste water remediation of a polyextremophilic α-amylase from an obligate halophilic Aspergillus gracilis. BioMed Res. Int. 2014, 2014, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batista-Garcia, R.A.; Balcazar-Lopez, E.; Miranda-Miranda, E.; Sanchez-Reyes, A.; Cuervo-Soto, L.; Aceves-Zamudio, D.; Atriztan-Hernandez, K.; Morales-Herrera, C.; Rodriguez-Hernandez, R.; Folch-Mallol, J. Characterization of lignocellulolytic activities from a moderate halophile strain of Aspergillus caesiellus isolated from a sugarcane bagasse fermentation. PLoS ONE 2014, 9, e105893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Annapurna, S.A.; Amarnath, S.; Shashank, G.; Anupam, K.; Kumar, H. Screening, isolation and characterization of protease producing moderately halophilic microorganisms. Asian J. Microbiol. Biotechnol. Environ. Sci. 2012, 14, 603–612. [Google Scholar]
- Yin, L.-J.; Hsu, T.-H.; Jiang, S.-T. Characterization of acidic protease from Aspergillus niger BCRC 32720. J. Agric. Food Chem. 2013, 61, 662–666. [Google Scholar] [CrossRef]
- de Castro, R.J.S.; Sato, H.H. Protease from Aspergillus oryzae: Biochemical characterization and application as a potential biocatalyst for production of protein hydrolysates with antioxidant activities. J. Food Process. 2014, 2014, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Souza, P.M.; Werneck, G.; Aliakbarian, B.; Siqueira, F.; Filho, E.X.F.; Perego, P.; Converti, A.; Magalhaes, P.O.; Pessoa, A. Production, purification and characterization of an aspartic protease from Aspergillus foetidus. Food Chem. Toxicol. 2017, 109, 1103–1110. [Google Scholar] [CrossRef]
- Monod, M.; Togni, G.; Rahalison, L.; Frenk, E. Isolation and characterization of an extracellular alkaline protease of Aspergillus fumigatus. J. Med. Microbiol. 1991, 35, 23–28. [Google Scholar] [CrossRef]
- Abu-Tahon, M.A.; Arafat, H.H.; Isaac, G.S. Laundry detergent compatibility and dehairing efficiency of alkaline thermostable protease produced from Aspergillus terreus under solid-state fermentation. J. Oleo Sci. 2020, 69, 241–254. [Google Scholar] [CrossRef] [Green Version]
- Solanki, P.; Putatunda, C.; Kumar, A.; Bhatia, R.; Walla, A. Microbial proteases: Ubiquitous enzymes with innumerable uses. 3 Biotech 2021, 11, 428. [Google Scholar] [CrossRef]
- Kamekura, M.; Onishi, H. Protease formation by a moderately halophilic Bacillus strain. Appl. Microbiol. 1974, 27, 809–810. [Google Scholar] [CrossRef] [PubMed]
- van Qua, D.; Simidu, U.; Taga, N. Purification and some properties of halophilic protease produced by a moderately halophilic marine Psuedomonas sp. Can. J. Microbiol. 1981, 27, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Karbalaei-Heidari, H.R.; Amoozegar, M.A.; Hajighasemi, M.; Ziaee, A.-A.; Ventosa, A. Production, optimization and purification of a novel extracellular protease from the moderately halophilic bacterium Halobacillus karajensis. J. Ind. Microbiol. Biotechnol. 2009, 36, 21–27. [Google Scholar] [CrossRef]
- Irimia, A.; Ebel, C.; Madern, D.; Richard, S.B.; Cosenza, L.W.; Zaccai, G.; Vellieux, F.M. The oligomeric states of Haloarcula marismortui malate dehydrogenase are modulated by solvent components as shown by crystallographic and biochemical studies. J. Mol. Biol. 2003, 3, 859–873. [Google Scholar] [CrossRef]
- Karen, R.; Capes, M.D.; DasSarma, S. Function and biotechnology of extremophilic enzymes in low water activity. Aqua. Biosyst. 2012, 8, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Riordan, J.F. The role of metals in enzyme activity. Ann. Clin. Lab. Sci. 1977, 7, 119–129. [Google Scholar]
- Sinha, R.; Khare, S.K. Characterization of detergent compatible protease of a halophilic Bacillus sp. EMB9: Differential role of metal ions in stability and activity. Bioresour. Technol. 2013, 145, 357–361. [Google Scholar] [CrossRef]
- Johnvesly, B.; Naik, G.R. Studies on production of thermostable alkaline protease from thermophilic and alkaliphilic Bacillus sp. JB-99 in a chemically defined medium. Process. Biochem. 2001, 37, 139–144. [Google Scholar] [CrossRef]
- Sayem, S.M.A.; Alam, M.J.; Hoq, M.M. Effect of temperature, pH, and metal ions on the activity and stability of alkaline protease from novel Bacillus licheniformis MZK03. Proc. Pak. Acad. Sci. 2006, 43, 257–262. [Google Scholar]
- Vidyasagar, M.; Prakash, S.; Litchfield, C.; Speeramulu, K. Purification and characterization of a thermostable, haloalkaliphilic extracellular serine protease from the extreme halophilic archaeon Halogeometricum borinquense strain TSS101. Archaea 2006, 2, 51–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sattar, H.; Aman, A.; Qader, S.A.U. Effect of metal ions, solvents and surfactants on the activity of protease from Aspergillus niger KIBGE-IB36. J. Basic Appl. Sci. 2017, 13, 491–495. [Google Scholar]
- Gupta, A.; Roy, I.; Patel, R.K.; Singh, S.P.; Khare, S.K.; Gupta, M.N. One-step purification and characterization of an alkaline protease from haloalkaliphilic Bacillus sp. J. Chromatogr. A 2005, 1075, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Matkawala, F.; Nighojkar, S.; Kumar, A.; Nighojkar, A. Microbial alkaline serine proteases: Production, properties and applications. World J. Microbiol. Biotechnol. 2021, 37, 63. [Google Scholar] [CrossRef] [PubMed]






| Metal Ion Concentration | ||
|---|---|---|
| Compounds | 2 mM | 4 mM |
| CaCl2 | 85.5 ± 6.9% | 71.8 ± 5.6% |
| MgSO4 | 91.1 ± 4.6% | 88.1 ± 5.0% |
| ZnSO4 | 59.7 ± 4.3% | 40.8 ± 7.2% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chung, D.; Yu, W.-J.; Lim, J.-Y.; Kang, N.-S.; Kwon, Y.-M.; Choi, G.; Bae, S.-S.; Cho, K.; Lee, D.-S. Characterization of the Proteolytic Activity of a Halophilic Aspergillus reticulatus Strain SK1-1 Isolated from a Solar Saltern. Microorganisms 2022, 10, 29. https://doi.org/10.3390/microorganisms10010029
Chung D, Yu W-J, Lim J-Y, Kang N-S, Kwon Y-M, Choi G, Bae S-S, Cho K, Lee D-S. Characterization of the Proteolytic Activity of a Halophilic Aspergillus reticulatus Strain SK1-1 Isolated from a Solar Saltern. Microorganisms. 2022; 10(1):29. https://doi.org/10.3390/microorganisms10010029
Chicago/Turabian StyleChung, Dawoon, Woon-Jong Yu, Ji-Yeon Lim, Nam-Seon Kang, Yong-Min Kwon, Grace Choi, Seung-Sub Bae, Kichul Cho, and Dae-Sung Lee. 2022. "Characterization of the Proteolytic Activity of a Halophilic Aspergillus reticulatus Strain SK1-1 Isolated from a Solar Saltern" Microorganisms 10, no. 1: 29. https://doi.org/10.3390/microorganisms10010029
APA StyleChung, D., Yu, W. -J., Lim, J. -Y., Kang, N. -S., Kwon, Y. -M., Choi, G., Bae, S. -S., Cho, K., & Lee, D. -S. (2022). Characterization of the Proteolytic Activity of a Halophilic Aspergillus reticulatus Strain SK1-1 Isolated from a Solar Saltern. Microorganisms, 10(1), 29. https://doi.org/10.3390/microorganisms10010029