Characterization of the 3,4-Dichloroaniline Degradation Gene Cluster in Acinetobacter soli GFJ2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Culture Conditions, and DNA Manipulations
2.2. Genome Sequence Analysis
2.3. Construction of Disruption Mutants
2.4. Reverse Transcription PCR and Quantitative Reverse Transcription PCR
2.5. Expression of dcd Genes in E. coli
2.6. Determination of Substrate Residue (34DCA)
2.7. Accession Numbers
2.8. Statistical Analysis
3. Results
3.1. Determination of the Genome Sequence of Strain GFJ2
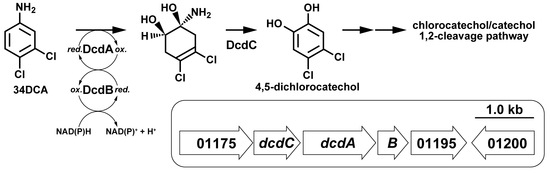
3.2. Identification of 34DCA-Degrading Genes
3.3. Transcriptional Induction of the dcd Gene Cluster
3.4. Disruption of dcdA and dcdB in GFJ2
3.5. Determination of the Roles of the dcdA, dcdB, and dcdC Genes in 34DCA Degradation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tran, A.T.; Hyne, R.V.; Doble, P. Determination of commonly used polar herbicides in agricultural drainage waters in Australia by HPLC. Chemosphere 2007, 67, 944–953. [Google Scholar] [CrossRef]
- Hussain, S.; Arshad, M.; Springael, D.; Sørensen, S.R.; Bending, G.; Devers-Lamrani, M.; Maqbool, Z.; Martin-Laurent, F. Abiotic and biotic processes governing the fate of phenylurea herbicides in soils: A review. Crit. Rev. Environ. Sci. Technol. 2015, 45, 1947–1998. [Google Scholar] [CrossRef]
- Burant, A.; Selbig, W.; Furlong, E.T.; Higgins, C.P. Trace organic contaminants in urban runoff: Associations with urban land-use. Environ. Pollut. 2018, 242, 2068–2077. [Google Scholar] [CrossRef]
- Boscolo, C.N.P.; Pereira, T.S.B.; Batalhao, I.G.; Dourado, P.L.R.; Schlenk, D.; de Almeida, E.A. Diuron metabolites act as endocrine disruptors and alter aggressive behavior in Nile tilapia (Oreochromis niloticus). Chemosphere 2018, 191, 832–838. [Google Scholar] [CrossRef]
- Alba, L.M.; Esmeralda, M.; Jaime, V. Enhanced biodegradation of phenylurea herbicides by Ochrobactrum anthrophi CD3 assessment of its feasibility in diuron-contaminated soils. Int. J. Environ. Res. Public Health 2022, 19, 1365. [Google Scholar] [CrossRef] [PubMed]
- Guzzella, L.; Capri, E.; Di Corcia, A.; Barra Caracciolo, A.; Giuliano, G. Fate of diuron and linuron in a field lysimeter experiment. J. Environ. Qual. 2006, 35, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Dejonghe, W.; Goris, J.; Dierickx, A.; De Dobbeleer, V.; Crul, K.; De Vos, P.; Verstraete, W.; Top, E.M. Diversity of 3-chloroaniline and 3,4-dichloroaniline degrading bacteria isolated from three different soils and involvement of their plasmids in chloroaniline degradation. FEMS Microbiol. Ecol. 2002, 42, 315–325. [Google Scholar] [CrossRef]
- Zhang, J.; Sun, J.Q.; Yuan, Q.Y.; Li, C.; Yan, X.; Hong, Q.; Li, S.P. Characterization of the propanil biodegradation pathway in Sphingomonas sp. Y57 and cloning of the propanil hydrolase gene prpH. J. Hazard. Mater. 2011, 196, 412–419. [Google Scholar] [CrossRef]
- Sorensen, S.R.; Rasmussen, J.; Jacobsen, C.S.; Jacobsen, O.S.; Juhler, R.K.; Aamand, J. Elucidating the key member of a linuron-mineralizing bacterial community by PCR and reverse transcription-PCR denaturing gradient gel electrophoresis 16S rRNA gene fingerprinting and cultivation. Appl. Environ. Microbiol. 2005, 71, 4144–4148. [Google Scholar] [CrossRef]
- Horemans, B.; Bers, K.; Ruiz Romero, E.; Pose Juan, E.; Dunon, V.; De Mot, R.; Springael, D. Functional redundancy of linuron degradation in microbial communities in agricultural soil and biopurification systems. Appl. Environ. Microbiol. 2016, 82, 2843–2853. [Google Scholar] [CrossRef]
- Bers, K.; Leroy, B.; Breugelmans, P.; Albers, P.; Lavigne, R.; Sorensen, S.R.; Aamand, J.; De Mot, R.; Wattiez, R.; Springael, D. A novel hydrolase identified by genomic-proteomic analysis of phenylurea herbicide mineralization by Variovorax sp. strain SRS16. Appl. Environ. Microbiol. 2011, 77, 8754–8764. [Google Scholar] [CrossRef]
- Nitisakulkan, T.; Oku, S.; Kudo, D.; Nakashimada, Y.; Tajima, T.; Vangnai, A.S.; Kato, J. Degradation of chloroanilines by toluene dioxygenase from Pseudomonas putida T57. J. Biosci. Bioeng. 2014, 117, 292–297. [Google Scholar] [CrossRef]
- Yao, X.F.; Khan, F.; Pandey, R.; Pandey, J.; Mourant, R.G.; Jain, R.K.; Guo, J.H.; Russell, R.J.; Oakeshott, J.G.; Pandey, G. Degradation of dichloroaniline isomers by a newly isolated strain, Bacillus megaterium IMT21. Microbiology (Reading) 2011, 157, 721–726. [Google Scholar] [CrossRef]
- Arora, P.K. Bacterial degradation of monocyclic aromatic amines. Front. Microbiol. 2015, 6, 820. [Google Scholar] [CrossRef]
- Hongsawat, P.; Vangnai, A.S. Biodegradation pathways of chloroanilines by Acinetobacter baylyi strain GFJ2. J. Hazard. Mater. 2011, 186, 1300–1307. [Google Scholar] [CrossRef]
- Araki, N.; Niikura, Y.; Miyauchi, K.; Kasai, D.; Masai, E.; Fukuda, M. Glucose-mediated transcriptional repression of PCB/biphenyl catabolic genes in Rhodococcus jostii RHA1. J. Mol. Microb. Biotech. 2011, 20, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Masai, E.; Yamada, A.; Healy, J.M.; Hatta, T.; Kimbara, K.; Fukuda, M.; Yano, K. Characterization of biphenyl catabolic genes of gram-positive polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 1995, 61, 2079–2085. [Google Scholar] [CrossRef]
- Kasai, D.; Fujinami, T.; Abe, T.; Mase, K.; Katayama, Y.; Fukuda, M.; Masai, E. Uncovering the protocatechuate 2,3-cleavage pathway genes. J. Bacteriol. 2009, 191, 6758–6768. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Lagesen, K.; Hallin, P.; Rodland, E.A.; Staerfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef] [PubMed]
- Sharp, J.O.; Sales, C.M.; LeBlanc, J.C.; Liu, J.; Wood, T.K.; Eltis, L.D.; Mohn, W.W.; Alvarez-Cohen, L. An inducible propane monooxygenase is responsible for N-nitrosodimethylamine degradation by Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 2007, 73, 6930–6938. [Google Scholar] [CrossRef] [PubMed]
- Van der Geize, R.; Hessels, G.I.; van Gerwen, R.; van der Meijden, P.; Dijkhuizen, L. Unmarked gene deletion mutagenesis of kstD, encoding 3-ketosteroid Δ1-dehydrogenase, in Rhodococcus erythropolis SQ1 using sacB as counter-selectable marker. FEMS Microbiol. Lett. 2001, 205, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, E.R.; Hara, H.; Miyazawa, D.; Davies, J.E.; Eltis, L.D.; Mohn, W.W. Transcriptomic assessment of isozymes in the biphenyl pathway of Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 2006, 72, 6183–6693. [Google Scholar] [CrossRef] [PubMed]
- Kasai, D.; Imai, S.; Asano, S.; Tabata, M.; Iijima, S.; Kamimura, N.; Masai, E.; Fukuda, M. Identification of natural rubber degradation gene in Rhizobacter gummiphilus NS21. Biosci. Biotechnol. Biochem. 2017, 81, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Sadauskas, M.; Vaitekunas, J.; Gasparaviciute, R.; Meskys, R. Indole biodegradation in Acinetobacter sp. strain O153: Genetic and biochemical characterization. Appl. Environ. Microbiol. 2017, 83, e01453-17. [Google Scholar] [CrossRef]
- Rabuffetti, M.; Cannazza, P.; Contente, M.L.; Pinto, A.; Romano, D.; Hoyos, P.; Alcantara, A.R.; Eberini, I.; Laurenzi, T.; Gourlay, L.; et al. Structural insights into the desymmetrization of bulky 1,2-dicarbonyls through enzymatic monoreduction. Bioorg. Chem. 2021, 108, 104644. [Google Scholar] [CrossRef]
- You, I.S.; Bartha, R. Stimulation of 3,4-dichloroaniline mineralization by aniline. Appl. Environ. Microbiol. 1982, 44, 678–681. [Google Scholar] [CrossRef]
- Lin, G.H.; Chen, H.P.; Shu, H.Y. Detoxification of indole by an indole-induced flavoprotein oxygenase from Acinetobacter baumannii. PLoS ONE 2015, 10, e0138798. [Google Scholar] [CrossRef] [PubMed]
- Gallegos, M.T.; Schleif, R.; Bairoch, A.; Hofmann, K.; Ramos, J.L. AraC/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 1997, 61, 393–410. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Huo, L.; Schleif, R. Repression of the araBAD promoter from araO1. J. Mol. Biol. 1992, 224, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Dunn, T.M.; Hahn, S.; Ogden, S.; Schleif, R.F. An operator at -280 base pairs that is required for repression of araBAD operon promoter: Addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc. Natl. Acad. Sci. USA 1984, 81, 5017–5020. [Google Scholar] [CrossRef] [PubMed]
- Miyada, C.G.; Stoltzfus, L.; Wilcox, G. Regulation of the araC gene of Escherichia coli: Catabolite repression, autoregulation, and effect on araBAD expression. Proc. Natl. Acad. Sci. USA 1984, 81, 4120–4124. [Google Scholar] [CrossRef]





| Gene | Deduced Molecular Mass (Da) a | Representative Homolog | Identity (%) b | Accession No. |
|---|---|---|---|---|
| BEN76_01175 | 45,749 (415) | Dienelactone hydrolase family protein (IifA) from Acinetobacter sp. | 89 | ARO76326 |
| Dienelactone hydrolase from Azospirillum brasilense | 22 | Q43914 | ||
| Dienelactone hydrolase from Pseudomonas putida | 20 | P0A114 | ||
| BEN76_01180 (dcdC) | 28,223 (263) | Short-chain dehydrogenase (IifB) from Acinetobacter sp. | 81 | ARO76327 |
| Dihydroanticapsin 7-dehydrogenase (BacC) from Bacillus subtilis | 30 | P39640 | ||
| 3-Oxoacyl-[acyl-carrier-protein] reductase (FabG) from Aquifex aeolicus | 30 | O67610 | ||
| BEN76_01185 (dcdA) | 46,014 (412) | Indole dioxygenase (IifC) from Acinetobacter sp. | 87 | ARO76328 |
| Styrene monooxygenase (StyA) from Pseudomonas sp. | 29 | O50214 | ||
| BEN76_01190 (dcdB) | 19,036 (172) | Flavin reductase (IifD) from Acinetobacter sp. | 90 | ARO76329 |
| Reductase component (HsaB) of 3-hydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione 4-hydroxylase from Rhodococcus jostii | 31 | Q0S808 | ||
| Reductase component (NtaB) of p-hydroxyphenylacetate 3-hydroxylase from Acinetobacter baumannii | 22 | Q6Q271 | ||
| BEN76_01195 | 34,850 (315) | MetA-pathway of phenol degradation (IifE) from Acinetobacter sp. | 83 | ARO76330 |
| Hypothetical protein from Acinetobacter baumannii | 80 | ENW75085 | ||
| BEN76_01200 | 40,833 (353) | AraC-type transcriptional regulator (IifR) from A. baumannii | 73 | F911_02001 |
| AraC/XylS-type transcriptional regulator (AntR) from Burkholderia cepacian | 23 | Q84BZ4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gibu, N.; Kasai, D.; Sato, S.; Tabata, M.; Vangnai, A.; Fukuda, M. Characterization of the 3,4-Dichloroaniline Degradation Gene Cluster in Acinetobacter soli GFJ2. Microorganisms 2024, 12, 613. https://doi.org/10.3390/microorganisms12030613
Gibu N, Kasai D, Sato S, Tabata M, Vangnai A, Fukuda M. Characterization of the 3,4-Dichloroaniline Degradation Gene Cluster in Acinetobacter soli GFJ2. Microorganisms. 2024; 12(3):613. https://doi.org/10.3390/microorganisms12030613
Chicago/Turabian StyleGibu, Namiko, Daisuke Kasai, Saki Sato, Michiro Tabata, Alisa Vangnai, and Masao Fukuda. 2024. "Characterization of the 3,4-Dichloroaniline Degradation Gene Cluster in Acinetobacter soli GFJ2" Microorganisms 12, no. 3: 613. https://doi.org/10.3390/microorganisms12030613
APA StyleGibu, N., Kasai, D., Sato, S., Tabata, M., Vangnai, A., & Fukuda, M. (2024). Characterization of the 3,4-Dichloroaniline Degradation Gene Cluster in Acinetobacter soli GFJ2. Microorganisms, 12(3), 613. https://doi.org/10.3390/microorganisms12030613