Microbial Biofilms and Chronic Wounds
Abstract
:1. Introduction—Microbial Biofilms
2. Biofilm Formation
3. Quorum Sensing
4. Biofilm Resistance
5. Biofilm Structure and Morphology
6. Biofilms in Chronic Wounds
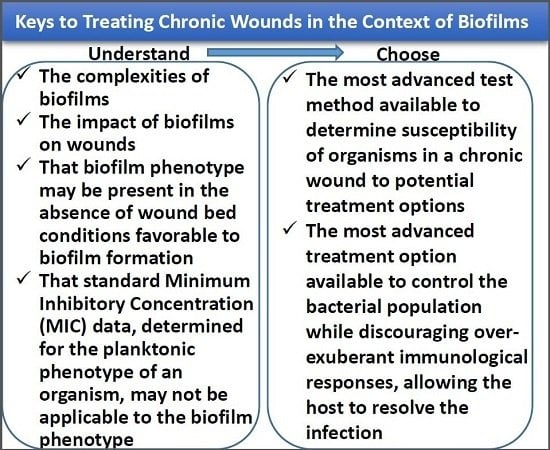
7. Identification of Drug Resistance/Tolerance, Susceptibility, and Treatments
8. Research on New Treatment Strategies
- Signals and cues:
- AIP-1 (Gram-positive bacteria)—controls the agr system based on quorum sensing such that disassembly of the biofilm is coupled to increased density or decreased nutrient availability, causing release of proteases and pore-forming toxins, and increased expression of virulence factors [100,104,105,106]
- Cell envelope-modifying molecules:
9. Summary
Acknowledgments
Conflicts of Interest
References
- Costerton, J.W.; Cheng, K.J.; Geesey, G.G.; Ladd, T.I.; Nickel, J.C.; Dasgupta, M.; Marrie, T.J. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 1987, 41, 435–464. [Google Scholar] [CrossRef] [PubMed]
- Cowan, T. Biofilms and their management: From concept to clinical reality. J. Wound Care 2011, 20, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Branda, S.S.; Vik, S.; Friedman, L.; Kolter, R. Biofilms: The matrix revisited. Trends Microbiol. 2005, 13, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Chapman, M.R.; Robinson, L.S.; Pinkner, J.S.; Roth, R.; Heuser, J.; Hammar, M.; Normark, S.; Hultgren, S.J. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 2002, 295, 851–855. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, N.; Kolodkin-Gal, I. The matrix reloaded: Probing the extracellular matrix synchronizes bacterial communities. J. Bacteriol. 2015, 197, 2092–2103. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, P.; Maira-Litran, T.; McBain, A.J.; Rickard, A.H.; Whyte, F.W. The physiology and collective recalcitrance of microbial biofilm communities. Adv. Microb. Physiol. 2002, 46, 202–256. [Google Scholar] [PubMed]
- Wilking, J.N.; Zaburdaev, V.; de Volder, M.; Losick, R.; Brenner, M.P.; Weitz, D.A. Liquid transport facilitated by channels in Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. USA 2013, 110, 848–852. [Google Scholar] [CrossRef] [PubMed]
- Lejeune, P. Contamination of abiotic surfaces: What a colonizing bacterium sees and how to blur it. Trends Microbiol. 2003, 11, 179–184. [Google Scholar] [CrossRef]
- Friedman, L.; Kolter, R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 2004, 51, 675–690. [Google Scholar] [CrossRef] [PubMed]
- Busscher, H.J.; van der Mei, H.C. Physico-chemical interactions in initial microbial adhesion and relevance for biofilm formation. Adv. Dent. Res. 1997, 11, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Aspiras, M.B.; Kazmerzak, K.M.; Kolenbrander, P.E.; McNab, R.; Hardegen, N.; Jenkinson, H.F. Expression of green fluorescent protein in Streptococcus gordonii DL1 and its use as a species-specific marker in coadhesion with Streptococcus oralis 34 in saliva-conditioned biofilms in vitro. Appl. Environ. Microbiol. 2000, 66, 4074–4083. [Google Scholar] [CrossRef] [PubMed]
- Rendueles, O.; Ghigo, J.-M. Multi-species biofilms: How to avoid unfriendly neighbors. FEMS Microbiol. Rev. 2012, 36, 972–989. [Google Scholar] [CrossRef] [PubMed]
- Diggle, S.P.; Winzer, K.; Lazdunski, A.; Williams, P.; Cámara, M. Advancing the quorum in Pseudomonas aeruginosa: MvaT and the regulation of N-acylhomoserine lactone production and virulence gene expression. J. Bacteriol. 2002, 184, 2576–2586. [Google Scholar] [CrossRef] [PubMed]
- Antunes, L.C.; Ferreira, R.B. Intercellular communication in bacteria. Crit. Rev. Microbiol. 2009, 35, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Kleerebezem, M.; Quadri, L.E.; Kuipers, O.P.; de Vos, W.M. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol. Microbiol. 1997, 24, 895–904. [Google Scholar] [CrossRef] [PubMed]
- Novick, R.P.; Geisinger, E. Quorum sensing in staphylococci. Annu. Rev. Genet. 2008, 42, 541–564. [Google Scholar] [CrossRef] [PubMed]
- Kong, K.F.; Vuong, C.; Otto, M. Staphylococcus quorum sensing in biofilm formation and infection. Int. J. Med. Microbiol. 2006, 296, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Vuong, C.; Saenz, H.L.; Gotz, F.; Otto, M. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 2000, 182, 1688–1693. [Google Scholar] [CrossRef] [PubMed]
- Lyczak, J.B.; Cannon, C.L.; Pier, G.B. Establishment of Pseudomonas aeruginosa infection: Lessons from a versatile opportunist. Microbes Infect. 2000, 2, 1051–1060. [Google Scholar] [CrossRef]
- Davies, D.G.; Parsek, M.R.; Pearson, J.P.; Iglewski, B.H.; Costerton, J.W.; Greenberg, E.P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 1998, 280, 295–298. [Google Scholar] [CrossRef] [PubMed]
- Erickson, D.L.; Endersby, R.; Kirkham, A.; Stuber, K.; Vollman, D.D.; Rabin, H.R.; Mitchell, I.; Storey, D.G. Pseudomonas aeruginosa quorum-sensing systems may control virulence factor expression in the lungs of patients with cystic fibrosis. Infect. Immun. 2002, 70, 1783–1790. [Google Scholar] [CrossRef] [PubMed]
- Karatuna, O.; Yagci, A. Analysis of the quorum sensing-dependent virulence factor production and its relationship with antimicrobial susceptibility in Pseudomonas aeruginosa respiratory isolates. Clin. Microbiol. Infect. 2010, 16, 1770–1775. [Google Scholar] [CrossRef] [PubMed]
- Heurlier, K.; Denervaud, V.; Haas, D. Impact of quorum sensing on fitness of Pseudomonas aeruginosa. Int. J. Med. Microbiol. 2006, 296, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Darch, S.E.; West, S.A.; Winzer, K.; Diggle, S.P. Density-dependent fitness benefits in quorum-sensing bacterial populations. Proc. Natl. Acad. Sci. USA 2012, 109, 8259–8263. [Google Scholar] [CrossRef] [PubMed]
- Skindersoe, M.E.; Zeuthen, L.H.; Brix, S.; Fink, L.N.; Lazenby, J.; Whittall, C.; Williams, P.; Diggle, S.P.; Froekiaer, H.; Cooley, M.; Givskov, M. Pseudomonas aeruginosa quorum-sensing signal molecules interfere with dendritic cell-induced T-cell proliferation. FEMS Immunol. Med. Microbiol. 2009, 55, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Reid, G. Regulatory and clinical aspects of dairy probiotics. In Report of a Joint FAO/WHO Expert Consultation on Evaluation of Health and Nutritional Properties of Probiotics in Food Including Powder Milk with Lactic Acid Bacteria; Food and Agricultural Organization of the United Nations: New York, NY, USA, 2001. [Google Scholar]
- White, C.; Sharman, A.K.; Gadd, G.M. An integrated microbial process for the bioremediation of soil contaminated with toxic metals. Nat. Biotechnol. 1998, 16, 572–575. [Google Scholar] [CrossRef] [PubMed]
- Paul, B.J.; Duthie, H.C.; Taylor, W.D. Nutrient cycling by biofilms in running waters of differing phosphorus status. J. North Am. Benthol. Soc. 1991, 10, 31–41. [Google Scholar] [CrossRef]
- Gilbert, P.; McBain, A.J. Biofilms: Their impact on health and their recalcitrance toward biocides. Am. J. Infect. Control. 2001, 29, 252–255. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.; Linton, S.; Corum, L.; Slone, W.; Okel, T.; Percival, S.L. The affect [sic] of pH and bacterial phenotypic state on antibiotic efficacy. Int. Wound J. 2012, 9, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Bayer, A.S.; Speert, D.P.; Park, S.; Tu, J.; Witt, M.; Nast, C.C.; Norman, D.C. Functional role of mucoid exopolysaccharide (alginate) in antibiotic-induced and polymorphonuclear leukocyte mediated killing of Pseudomonas aeruginosa. Infect. Immun. 1991, 59, 302–308. [Google Scholar] [PubMed]
- Thurlow, L.R.; Hanke, M.L.; Fritz, T.; Angle, A.; Aldrich, A.; Williams, S.H.; Engebretsen, I.L.; Bayles, K.W.; Horswill, A.R.; Kielian, T. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J. Immunol. 2011, 186, 6585–6596. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M. Bacterial biofilms and human disease. Sci. Prog. 2001, 84, 235–254. [Google Scholar] [CrossRef] [PubMed]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef] [PubMed]
- Malic, S.; Hill, K.E.; Playle, R.; Thomas, D.W.; Williams, D.W. In vitro interactions of chronic wound bacteria in biofilms. J. Wound Care. 2011, 20, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, J.B. Antibiotic-induced biofilm formation. Int. J. Artif. Organs. 2011, 34, 737–751. [Google Scholar] [CrossRef] [PubMed]
- Weiser, J.; Henke, H.A.; Hector, N.; Both, A.; Christner, M.; Buttner, H.; Kaplan, J.B.; Rohde, H. Sub-inhibitory tigecycline concentrations induce extracellular matrix binding protein Embp dependent Staphylococcus epidermidis biofilm formation and immune evasion. Int. J. Med. Microbiol. 2016, 306, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Mlynek, K.D.; Callahan, M.T.; Shimkevitch, A.V.; Farmer, J.T.; Endres, J.L.; Marchand, M.; Bayles, K.W.; Horswill, A.R.; Kaplan, J.B. Effects of low-dose amoxicillin on Staphylococcus aureus USA300 biofilms. Antimicrob. Agents Chemother. 2016, 60, 2639–2651. [Google Scholar] [CrossRef] [PubMed]
- Habash, M.B.; van der Mei, H.C.; Busscher, H.J.; Reid, G. The effect of water, ascorbic acid, and cranberry derived supplementation on human urine and uropathogen adhesion to silicone rubber. Can. J. Microbiol. 1999, 45, 691–694. [Google Scholar] [CrossRef] [PubMed]
- Wolcott, R.D.; Rhoads, D.D.; Bennett, M.E.; Wolcott, B.M.; Gogokhia, L.; Costerton, J.W.; Dowd, S.E. Chronic wounds and the medical biofilm paradigm. J. Wound Care. 2010, 19, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Govan, J.R.W.; Deretic, V. Microbial pathogenesis in cystic fibrosis: Mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 1996, 60, 539–574. [Google Scholar] [PubMed]
- Minutes of the National Advisory Dental and Craniofacial Research Council. In Presented at 153rd Meeting, Bethesda, MD, USA, September 1997.
- Costerton, J.W.; Lewandowski, Z.; Caldwell, D.E.; Korber, D.R.; Lappin-Scott, H.M. Microbial biofilms. Annu. Rev. Microbiol. 1995, 49, 711–745. [Google Scholar] [CrossRef] [PubMed]
- Wolcott, R.D.; Rhoads, D.D.; Dowd, S.E. Biofilms and chronic wound inflammation. J. Wound Care. 2008, 17, 333–341. [Google Scholar] [CrossRef] [PubMed]
- McBain, A.J.; Allison, D.G.; Gilbert, P. Emerging strategies for the chemical treatment of microbial biofilms. Biotechnol. Genet. Eng. Rev. 2000, 17, 267–279. [Google Scholar] [CrossRef] [PubMed]
- Fux, C.A.; Costerton, J.W.; Stewart, P.S.; Stoodley, P. Survival strategies of infectious biofilms. TRENDS Microbiol. 2005, 13, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Persister cells and the riddle of biofilm survival. Biochemistry (Mosc.) 2005, 70, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Percival, S.L.; Hill, K.E.; Malic, S.; Thomas, D.W.; Williams, D.W. Antimicrobial tolerance and the significance of persister cells in recalcitrant chronic wound biofilms. Wound Repair Regen. 2011, 19, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Percival, S.L.; Emanuel, C.; Cutting, K.F.; Williams, D.W. Microbiology of the skin and the role of biofilms in infection. Int. Wound J. 2012, 9, 14–32. [Google Scholar] [CrossRef] [PubMed]
- Borges-Walmsley, M.I.; McKeegan, K.S.; Walmsley, A.R. Structure and function of efflux pumps that confer resistance to drugs. Biochem. J. 2003, 376, 313–338. [Google Scholar] [CrossRef] [PubMed]
- Mah, T.F.; Pitts, B.; Pellock, B.; Walker, G.C.; Stewart, P.S.; O’Toole, G.A. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 2003, 426, 306–310. [Google Scholar] [CrossRef] [PubMed]
- Hall-Stoodley, L.; Stoodley, P. Developmental regulation of microbial biofilms. Curr. Opin. Biotech. 2002, 13, 228–233. [Google Scholar] [CrossRef]
- Kjelleberg, S.; Molin, S. Is there a role for quorum sensing signals in bacterial biofilms? Curr. Opin. Microbiol. 2002, 5, 254–258. [Google Scholar] [CrossRef]
- Schaudinn, C.; Stoodley, P.; Kainovic, A.; O’Keeffe, T.; Costerton, W.; Robinson, D.; Baum, M.; Ehrlich, G.; Webster, P. Bacterial biofilms, other structures seen as mainstream concepts. Microbe 2007, 2, 231–237. [Google Scholar] [CrossRef]
- Kempes, C.P.; Okegbe, C.; Mears-Clarke, Z.; Follows, M.J.; Deitrich, L.E.P. Morphological optimization for access to dual oxidants in biofilms. Proc. Natl. Acad. Sci. USA 2014, 111, 208–213. [Google Scholar] [CrossRef] [PubMed]
- Deitrich, L.E.P.; Okegbe, C.; Price-Whelan, A.; Sakhtah, H.; Hunter, R.C.; Newman, D.K. Bacterial community morphogenesis is intimately linked to the intracellular redox state. J. Bacteriol. 2013, 195, 1371–1380. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kern, S.E.; Newman, D.K. Endogenous phenazine antibiotics promote anaerobic survival of Pseudomonas aeruginosa via extracellular electron transfer. J. Bacteriol. 2010, 192, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Price-Whelan, A.; Dietrich, L.E.P.; Newman, D.K. Pyocyanin alters redox homeostatis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. J. Bacteriol. 2007, 189, 6372–6381. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, L.E.P.; Teal, T.K.; Price-Whelan, A.; Newman, D.K. Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science 2008, 321, 1203–1206. [Google Scholar] [CrossRef] [PubMed]
- Ramos, I.; Dietrich, L.E.P.; Price-Whelan, A.; Newman, D.K. Phenazines affect biofilm formation by Pseudomonas aeruginosa in similar ways at various scales. Res. Microbiol. 2010, 161, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Xavier, J.B.; Foster, K.R. Cooperation and conflict in microbial biofilms. Proc. Natl. Acad. Sci. USA 2007, 104, 876–881. [Google Scholar] [CrossRef] [PubMed]
- Nadell, C.D.; Foster, K.R.; Xavier, J.B. Emergence of spatial structure in cell groups and the evolution of cooperation. PLOS Comput. Biol. 2010, 6, e1000716. [Google Scholar] [CrossRef]
- Asally, M.; Kittisopikul, M.; Rue, P.; Du, Y.; Hu, Z.; Cagatay, T.; Robinson, A.B.; Lu, H.; Garcia-Ojalvo, J.; Suel, G.M. Localized cell death focuses mechanical forces during 3D patterning in a biofilm. Proc. Natl. Acad. Sci. USA 2012, 109, 18891–18896. [Google Scholar] [CrossRef] [PubMed]
- Kolodkin-Gal, I.; Elshotz, A.K.; Muth, C.; Girguis, P.R.; Kolter, R.; Losick, R. Respiration control of multicellularity in Bacillus subtilis by a complex of the cytochrome chain with a membrane-embedded histidine kinase. Genes Dev. 2013, 27, 887–899. [Google Scholar] [CrossRef] [PubMed]
- Oppenheimer-Shaanan, Y.; Sibony-Nevo, O.; Bloom-Ackermann, Z.; Suissa, R.; Steinberg, N.; Kartvelishvily, E.; Brumfeld, V.; Kolodkin-Gal, I. Spatio-temporal assembly of functional mineral scaffolds within microbial biofilms. Npj Biofilms. Microbiomes. 2016, 2, 15031. [Google Scholar] [CrossRef]
- Sauer, K.; Cullen, M.C.; Rickard, A.H.; Zeef, L.A.; Davies, D.G.; Gilbert, P. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J. Bacteriol. 2004, 186, 7312–7326. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.W.Y. Effect of Viscosity on Biofilm Phenotype Expression. MSc Thesis, Department of Biology, University of Calgary, Calgary, AB, Canada, 1999. [Google Scholar]
- Lam, K.; Olson, M.E.; Burrell, R.E.; Wright, J.B. Development of a porcine model for examining the influence of wound contamination/infection on wound healing. Wound Rep. Reg. 1999, 7, A305. [Google Scholar]
- Wright, J.B.; Olson, M.E.; Lam, K.; Burrell, R.E. Use of a porcine model to examine the impact of a silver-coated dressing on the rate of wound healing in contaminated full-thickness wounds. Wound Rep. Reg. 1999, 7, A326. [Google Scholar]
- Thomson, C.H. Biofilms: Do they affect wound healing? Int. Wound J. 2011, 8, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Roth, R.R.; James, W.D. Microbial ecology of the skin. Annu. Rev. Microbiol. 1988, 42, 441–464. [Google Scholar] [CrossRef] [PubMed]
- Krutmann, J. Pre- and probiotics for human skin. J. Dermatol. Sci. 2009, 54, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Gan, B.S.; Kim, J.; Reid, G.; Cadieux, P.; Howard, J.C. Lactobacillus fermentum RC-14 inhibits Staphylococcus aureus infection of surgical implants in rats. J. Infect. Dis. 2002, 185, 1369–1372. [Google Scholar] [CrossRef] [PubMed]
- Gough, E.; Shaikh, H.; Manges, A.R. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin. Infect. Dis. 2011, 53, 994–1002. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 2001, 45, 999–1007. [Google Scholar] [CrossRef] [PubMed]
- Olson, M.E.; Ceri, H.; Morck, D.W.; Buret, A.G.; Read, R.R. Biofilm bacteria: Formation and comparative susceptibility to antibiotics. Can. J. Vet. Res. 2002, 66, 86–92. [Google Scholar] [PubMed]
- Keren, I.; Kaldalu, N.; Spoering, A.; Wang, Y.; Lewis, K. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 2004, 230, 13–18. [Google Scholar] [CrossRef]
- Singh, R.; Ray, P.; Das, A.; Sharma, M. Role of persisters and small-colony variants in antibiotic resistance of planktonic and biofilm-associated Staphylococcus aureus: An in vitro study. J. Med. Microbiol. 2009, 58, 1067–1073. [Google Scholar] [CrossRef] [PubMed]
- Abraham, W.R. Controlling biofilms of gram-positive pathogenic bacteria. Curr. Med. Chem. 2006, 13, 1509–1524. [Google Scholar] [CrossRef] [PubMed]
- Kim, P.J.; Steinberg, J.S. Wound care: Biofilm and its impact on the latest treatment modalities for ulcerations of the diabetic foot. Semin. Vasc. Surg. 2012, 25, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Harrison-Balestra, C.; Cazzaniga, A.L.; Davis, S.C.; Mertz, P.M. A wound-isolated Pseudomonas aeruginosa grows a biofilm in vitro within 10 hours and is visualized by light microscopy. Dermatol. Surg. 2003, 29, 631–635. [Google Scholar] [PubMed]
- Wolcott, R.D.; Rumbaugh, K.P.; James, G.; Schultz, G.; Phillips, P.; Yang, Q.; Watters, C.; Steward, P.S.; Dowd, S.E. Biofilm maturity studies indicate sharp debridement opens a time-dependent therapeutic window. J. Wound Care. 2010, 19, 320–328. [Google Scholar] [CrossRef] [PubMed]
- James, G.A.; Swogger, E.; Wolcott, R.; Pulcini, E.; Secor, P.; Sestrich, J.; Costerton, J.W.; Stewart, P.S. Biofilms in chronic wounds. Wound Rep. Regen. 2008, 16, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Hurlow, J.; Bowler, P.J. Potential implications of biofilm in chronic wounds: A case series. J. Wound Care. 2012, 21, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Wright, J.B.; Lam, K.; Buret, A.G.; Olson, M.E.; Burrell, R.E. Early healing events in a porcine model of contaminated wounds: Impact of nanocrystalline silver on matrix metalloproteinases, cellular apoptosis and wound healing. Wound Repair Regen. 2002, 10, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Wright, J.B.; Lam, K.; Olson, M.E.; Burrell, R.E. Is antimicrobial efficacy sufficient? A question concerning the benefits of new dressings. Wounds 2003, 15, 133–142. [Google Scholar]
- Concepts in Care: Microbiology and Infectious Disease in Cystic Fibrosis; Saiman, L.; Schidlow, D.; Smith, A. (Eds.) Cystic Fibrosis Foundation: Bethesda, MD, USA, 1994; Volume V.
- Smith, A.L.; Fiel, S.B.; Mayer-Hamblett, N.; Ramsey, B.; Burns, J.L. Susceptibility testing of Pseudomonas aeruginosa isolates and clinical response to parental antibiotic administration: Lack of association in cystic fibrosis. Chest 2003, 123, 1495–1502. [Google Scholar] [CrossRef] [PubMed]
- Keays, T.; Ferris, W.; Vandemheen, K.L.; Chan, F.; Yeung, S.-W.; Mah, T.-F.; Ramotar, K.; Saginur, R.; Aaron, S.D. A retrospective analysis of biofilm antibiotic susceptibility testing: A better predictor of clinical response in cystic fibrosis exacerbations. J. Cyst. Fibros. 2009, 8, 122–127. [Google Scholar] [CrossRef] [PubMed]
- Ceri, H.; Olson, M.E.; Stremick, C.; Read, R.R.; Morck, D.; Buret, A. The Calgary biofilm device: New technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 1999, 37, 1771–1776. [Google Scholar] [PubMed]
- Hammond, A.A.; Miller, K.G.; Kruczek, C.J.; Dertien, J.; Colmer-Hamood, J.A.; Griswold, J.A.; Horswill, A.R.; Hamood, A.N. An in vitro biofilm model to examine the effect of antibiotic ointments on biofilms produced by burn wound bacterial isolates. Burns 2011, 37, 312–321. [Google Scholar] [CrossRef] [PubMed]
- ASTM International. Standard Test Method for Testing Disinfectant Efficacy against Pseudomonas aeruginosa Biofilm Using the MBEC™ Assay. E2799-12; ASTM International: West Conshohocken, PA, USA, 2012. [Google Scholar]
- Zhao, G.; Hochwalt, P.C.; Usui, M.L.; Underwood, R.A.; Singh, P.K.; James, G.A.; Stewart, P.S.; Fleckman, P.; Olerud, J.E. Delayed wound healing in diabetic (db/db) mice with Pseudomonas aeruginosa biofilm challenge: A model for the study of chronic wounds. Wound Repair Regen. 2010, 18, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Nadworny, P.; Burrell, R.E. A review of assessment techniques for silver technology in wound care, Part II: Tissue culture and in vivo methods for determining antimicrobial and anti-inflammatory activity. J. Wound Tech. 2008, 2, 14–22. [Google Scholar]
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E., Jr.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Coates, A.; Hu, Y. Novel approaches to developing new antibiotics for bacterial infections. Br. J. Pharmacol. 2007, 152, 1147–1154. [Google Scholar] [CrossRef] [PubMed]
- Rasko, D.A.; Sperandio, V. Anti-virulence strategies to combat bacteria-mediated disease. Nature Rev. Drug Discov. 2010, 9, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Maeda, T.; Garcia-Contreras, R.; Pu, M.; Sheng, L.; Garcia, L.R.; Tomas, M.; Wood, T.K. Quorum quenching quandary: Resistance to antivirulence compounds. ISME J. 2012, 6, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Romero, D.; Kolter, R. Will biofilm disassembly agents make it to market? Trends Microbiol. 2011, 19, 304–306. [Google Scholar] [CrossRef] [PubMed]
- Oppenheimer-Shaanan, Y.; Steinberg, N.; Kolodkin-Gal, I. Small molecules are natural triggers for the disassembly of biofilms. Trends Microbiol. 2013, 21, 594–601. [Google Scholar] [CrossRef] [PubMed]
- Karatan, E.; Watnick, P. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 2009, 73, 310–347. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, J.B. Biofilm dispersal: Mechanisms, clinical implications, and potential therapeutic uses. J. Dent. Res. 2010, 89, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Donlan, R.M. Preventing biofilms of clinically relevant organisms using bacteriophage. Trends Microbiol. 2009, 17, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Sakoulas, G.; Eliopoulos, G.M.; Moellering, R.C., Jr.; Novick, R.P.; Venkataraman, L.; Wennersten, C.; DeGirolami, P.C.; Schwaber, M.J.; Gold, H.S. Staphylococcus aureus accessory gene regulator (agr) group II: Is there a relationship to the development of intermediate-level glycopeptide resistance? J. Infect. Dis. 2003, 187, 929–938. [Google Scholar] [CrossRef] [PubMed]
- Vuong, C.; Durr, M.; Carmody, A.B.; Peschel, A.; Klebanoff, S.J.; Otto, M. Regulated expression of pathogen-associated molecular pattern molecules in Staphylococcus epidermidis: Quorum sensing determines pro-inflammatory capacity and production of phenol-soluble modulins. Cell. Microbiol. 2004, 6, 753–759. [Google Scholar] [CrossRef] [PubMed]
- Boles, B.R.; Horswill, A.R. agr-Mediated dispersal of Staphylococcus aureus biofilms. PLOS Pathogens. 2008, 4, e1000052. [Google Scholar] [CrossRef] [PubMed]
- Crossman, L.; Dow, J.M. Biofilm formation and dispersal in Xanthomonas campestris. Microbes. Infect. 2004, 6, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Davies, D.G.; Marques, C.N.H. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 2009, 191, 1393–1403. [Google Scholar] [CrossRef] [PubMed]
- Banin, E.; Vasil, M.L.; Greenberg, E.P. Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. USA 2005, 102, 11076–11081. [Google Scholar] [CrossRef] [PubMed]
- Cava, F.; de Pedro, M.A.; Lam, H.; Davis, B.M.; Waldor, M.K. Distinct pathways for modification of the bacterial cell wall by non-canonical d-amino acids. EMBO J. 2011, 30, 3442–3453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolodkin-Gal, I.; Romero, D.; Cao, S.; Clardy, J.; Kolter, R.; Losick, R. d-amino acids trigger biofilm disassembly. Science 2010, 328, 627–629. [Google Scholar] [CrossRef] [PubMed]
- Lopez, D.; Kolter, R. Functional microdomains in bacterial membranes. Genes Dev. 2010, 24, 1893–1902. [Google Scholar] [CrossRef] [PubMed]
- Yepes, A.; Schneider, J.; Mielich, B.; Koch, G.; Garcia-Betancur, J.C.; Ramamurthi, K.S.; Vlamakis, H.; Lopez, D. The biofilm formation defect of a Bacillus subtilis flotillin-defective mutant involves the protease FtsH. Mol. Microbiol. 2012, 86, 457–471. [Google Scholar] [CrossRef] [PubMed]
- Karatan, E.; Duncan, T.R.; Watnick, P.I. NspS, a predicted polyamine sensor, mediates activation of Vibrio cholerae biofilm formation by norspermidine. J. Bacteriol. 2005, 187, 7434–7443. [Google Scholar] [CrossRef] [PubMed]
- Parsek, M.R.; Greenberg, E.P. Sociomicrobiology: The connections between quorum sensing and biofilms. Trends Microbiol. 2005, 13, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W. The Biofilm Primer; Springer-Verlag Berlin Heidelberg: Berlin, Germany, 2007; p. 64. [Google Scholar]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Omar, A.; Wright, J.B.; Schultz, G.; Burrell, R.; Nadworny, P. Microbial Biofilms and Chronic Wounds. Microorganisms 2017, 5, 9. https://doi.org/10.3390/microorganisms5010009
Omar A, Wright JB, Schultz G, Burrell R, Nadworny P. Microbial Biofilms and Chronic Wounds. Microorganisms. 2017; 5(1):9. https://doi.org/10.3390/microorganisms5010009
Chicago/Turabian StyleOmar, Amin, J. Barry Wright, Gregory Schultz, Robert Burrell, and Patricia Nadworny. 2017. "Microbial Biofilms and Chronic Wounds" Microorganisms 5, no. 1: 9. https://doi.org/10.3390/microorganisms5010009
APA StyleOmar, A., Wright, J. B., Schultz, G., Burrell, R., & Nadworny, P. (2017). Microbial Biofilms and Chronic Wounds. Microorganisms, 5(1), 9. https://doi.org/10.3390/microorganisms5010009