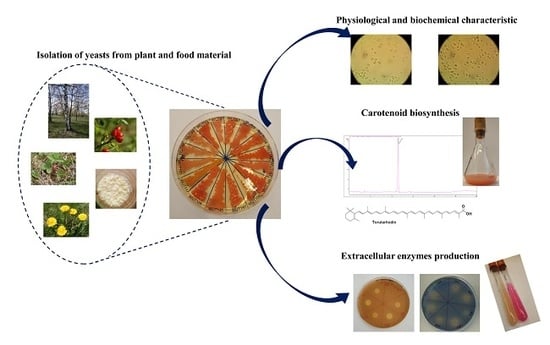
Carotenoid-Producing Yeasts: Identification and Characteristics of Environmental Isolates with a Valuable Extracellular Enzymatic Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolating the Yeast Strains
2.2. Polymerase Chain Reaction and the Sequencing of rDNA Fragments
2.3. Phylogenetic Analysis
2.4. Characterization of Isolated Yeast
2.5. Screening for Extracellular Enzymes Production
2.6. Determining Cellulolytic Activity in a Plate Assay and Liquid Assay
2.7. Carotenoids Production in Liquid Cultures
2.8. Isolation of Total Carotenoids and Lipids from Yeast Biomass
2.9. Dry Biomass Estimation
2.10. HPLC Analysis of Residual Sugars in Culture Media and Carotenoid Pigments Extracted from Yeast Biomass
3. Results and Discussion
3.1. Identification of Yeasts Isolated from Plant Samples
3.2. Physiological and Biochemical Characteristic of Yeasts
3.3. Synthesis of Cellulases by Selected Yeasts
3.4. Preliminary Evaluation of Carotenoid Production
3.4.1. First Test Group—Strains WUT10-WUT117
3.4.2. Second Test Group—strains WUT128-WUT194
3.5. The Effect of Lower Temperatures on Carotenoid Synthesis in a Batch Cultures of Cystobasidium Species
3.6. The Impact of Glucose Concentration on Pigment Accumulation in R. mucilaginosa and C. macerans Strains
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tsuji, M.; Tsujimoto, M.; Imura, S. Cystobasidium tubakii and Cystobasidium ongulense, new basidiomycetous yeast species isolated from East Ongul Island, East Antarctica. Mycoscience 2017, 58, 103–110. [Google Scholar] [CrossRef]
- Choudhary, D.K.; Bhavdish, N.J. Basidiomycetous Yeasts: Current Status. In Yeast Biotechnology: Diversity and Applications; Satyanarayana, T., Kunze, G., Eds.; Springer Nature: Dordrecht, The Netherlands, 2009; pp. 19–43. [Google Scholar]
- Tao, B.Y. Industrial Applications for Plant Oils and Lipids. Bioprocess. Value-Added Prod. Renew. Resour. 2007, 611–627. [Google Scholar] [CrossRef]
- Aksu, Z.; Eren, T.A. Carotenoids production by the yeast Rhodotorula mucilaginosa: Use of agricultural wastes as a carbon source. Process Biochem. 2005, 40, 2985–2991. [Google Scholar] [CrossRef]
- Tinoi, J.; Rakariyatham, N.; Deming, R.L. Simplex optimization of carotenoid production by Rhodotorula glutinis using hydrolyzed mung bean waste flour as substrate. Process Biochem. 2005, 40, 2551–2557. [Google Scholar] [CrossRef]
- Kot, A.M.; Błazejak, S.; Gientka, I.; Kieliszek, M.; Bryś, J. Torulene and torularhodin: ‘New’ fungal carotenoids for industry? Microb. Cell Fact. 2018, 17, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davoli, P.; Weber, R. Carotenoid pigments from the red mirror yeast, Sporobolomyces roseus. Mycologist 2002, 16, 102–108. [Google Scholar] [CrossRef]
- Hu, Z.C.; Zheng, Y.G.; Wang, Z.; Shen, Y.C. Effect of sugar-feeding strategies on astaxanthin production by Xanthophyllomyces dendrorhous. World J. Microbiol. Biotechnol. 2005, 21, 771–775. [Google Scholar] [CrossRef]
- Ramírez, J.; Obledo, N.; Arellano, M.; Herrera, E. Astaxanthin production by Phaffia rhodozyma in a fedbatch culture using a low cost medium feeding. e-Gnosis 2006, 4, 1–9. [Google Scholar]
- Ni, H.; Chen, Q.; He, G.; Wu, G.; Yang, Y. Optimization of acidic extraction of astaxanthin from Phaffia rhodozyma. J. Zhejiang Univ. Sci. B 2008, 9, 51–59. [Google Scholar] [CrossRef] [Green Version]
- Stolarzewicz, I.; Kapturowska, A.; Białecka-Florjańczyk, E. Mikrobiologiczne źródła barwników w technologii żywności. Postep. Mikrobiol. 2012, 51, 167–176. [Google Scholar]
- Barredo, J.L.; Garc, C.; Kosalkova, K.; Barreiro, C. Biosynthesis of astaxanthin as a main carotenoid in the Heterobasidiomycetous yeast Xanthophyllomyces dendrorhous. J. fungi 2017, 3, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mata-Gómez, L.C.; Montañez, J.C.; Méndez-Zavala, A.; Aguilar, C.N. Biotechnological production of carotenoids by yeasts: An overview. Microb. Cell Fact. 2014, 13, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stahl, W.; Sies, H. Carotenoids and protection against solar UV radiation. Skin Pharmacol. Appl. Skin Physiol. 2002, 15, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Fraser, P.D.; Bramley, P.M. The biosynthesis and nutritional uses of carotenoids. Prog. Lipid Res. 2004, 43, 228–265. [Google Scholar] [CrossRef] [PubMed]
- Ye, V.M.; Bhatia, S.K. Pathway engineering strategies for production of beneficial carotenoids in microbial hosts. Biotechnol. Lett. 2012, 34, 1405–1414. [Google Scholar] [CrossRef] [PubMed]
- Peralta, R.M.; da Silva, B.P.; Côrrea RC, G.; Kato, C.G.; Seixas FA, V.; Bracht, A. Enzymes from Basidiomycetes—Peculiar and Efficient Tools for Biotechnology. In Biotechnology of Microbial Enzymes; Brahmachari, G., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 119–149. [Google Scholar] [CrossRef]
- Mendonça Maciel, M.; Castro e Silva, A.; Camarão Telles Ribeiro, H. Industrial and biotechnological applications of ligninolytic enzymes of the basidiomycota: A review. Electron. J. Biotechnol. 2010, 13, 1–13. [Google Scholar] [CrossRef]
- Tanimura, A.; Sugita, T.; Endoh, R.; Ohkuma, M.; Kishino, S.; Ogawa, J.; Shima, J.; Takashima, M. Lipid production via simultaneous conversion of glucose and xylose by a novel yeast, Cystobasidium iriomotense. PLoS ONE 2018, 13, e0202164. [Google Scholar] [CrossRef] [Green Version]
- Vyas, S.; Chhabra, M. Isolation, identification and characterization of Cystobasidium oligophagum JRC1: A cellulase and lipase producing oleaginous yeast. Bioresour. Technol. 2017, 223, 250–258. [Google Scholar] [CrossRef]
- Rönnander, J.; Ljunggren, J.; Hedenström, E.; Wright, S.A.I. Biotransformation of vanillin into vanillyl alcohol by a novel strain of Cystobasidium laryngis isolated from decaying wood. AMB Express 2018, 8, 4–11. [Google Scholar] [CrossRef]
- Chreptowicz, K.; Sternicka, M.K.; Kowalska, P.D.; Mierzejewska, J. Screening of yeasts for the production of 2-phenylethanol (r ose aroma) in organic waste-based media. Lett. Appl. Microbiol. 2018, 66, 153–160. [Google Scholar] [CrossRef]
- Chreptowicz, K.; Wielechowska, M.; Główczyk-Zubek, J.; Rybak, E.; Mierzejewska, J. Production of natural 2-phenylethanol: From biotransformation to purified product. Food Bioprod. Process. 2016, 100, 275–281. [Google Scholar] [CrossRef]
- Adame-Soto, P.J.; Aréchiga-Carvajal, E.T.; López, M.G.; González-Herrera, S.M.; Moreno-Jiménez, M.R.; Urtiz-Estrada, N.; Rutiaga-Quiñones, O.M. Potential production of 2-phenylethanol and 2-phenylethylacetate by non-Saccharomyces yeasts from Agave durangensis. Ann. Microbiol. 2019, 69, 989–1000. [Google Scholar] [CrossRef]
- Ghose, T.K. Measurement of cellulase activities. Pure Appl. Chem. 1987, 59, 257–268. [Google Scholar] [CrossRef]
- Tkáčová, J.; Klempová, T.; Čertík, M. Kinetic study of growth, lipid and carotenoid formation in β-carotene producing Rhodotorula glutinis. Chem. Pap. 2018, 72, 1193–1203. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 2018, 35, 547–1549. [Google Scholar] [CrossRef] [PubMed]
- Coronado-Ruiz, C.; Avendaño, R.; Escudero-Leyva, E.; Conejo-Barboza, G.; Chaverri, P.; Chavarría, M. Two new cellulolytic fungal species isolated from a 19th-century art collection. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Zieniuk, B.; Fabiszewska, A. Yarrowia lipolytica: a beneficious yeast in biotechnology as a rare opportunistic fungal pathogen: a minireview. World J. Microbiol. Biotechnol. 2019, 35, 10. [Google Scholar] [CrossRef] [Green Version]
- Schlander, M.; Distler, U.; Tenzer, S.; Thines, E.; Claus, H. Purification and properties of yeast proteases secreted by Wickerhamomyces anomalus 227 and Metschnikovia pulcherrima 446 during growth in a white grape juice. Fermentation 2016, 3, 2. [Google Scholar] [CrossRef] [Green Version]
- Larios, A.; García, H.S.; Oliart, R.M.; Valerio-Alfaro, G. Synthesis of flavor and fragrance esters using Candida antarctica lipase. Appl. Microbiol. Biotechnol. 2004, 65, 373–376. [Google Scholar] [CrossRef]
- Taskin, M.; Ucar, M.H.; Unver, Y.; Kara, A.A.; Ozdemir, M.; Ortucu, S. Lipase production with free and immobilized cells of cold-adapted yeast Rhodotorula glutinis HL25. Biocatal. Agric. Biotechnol. 2016, 8, 97–103. [Google Scholar] [CrossRef]
- Papaparaskevas, D.; Christakopoulos, P.; Kekos, D.; Macris, B.J. Optimizing production of extracellular lipase from Rhodotorula glutinis. Biotechnol. Lett. 1992, 14, 397–402. [Google Scholar] [CrossRef]
- Weber, R.W.S.; Anke, H.; Davoli, P. Simple method for the extraction and reversed-phase high-performance liquid chromatographic analysis of carotenoid pigments from red yeasts (Basidiomycota, Fungi). J. Chromatogr. A 2007, 1145, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Smaniotto, A.; Skovronski, A.; Rigo, E.; Tsai, S.M.; Durrer, A.; Foltran, L.L.; di Luccio, M.; Oliveira, J.V.; de Oliveira, D.; Treichel, H. ‘Synthetic lipase’ production from a newly isolated Sporidiobolus pararoseus strain by submerged fermentation. Brazilian J. Microbiol. 2012, 43, 1490–1498. [Google Scholar] [CrossRef] [Green Version]
- Daskaya-Dikmen, C.; Karbancioglu-Guler, F.; Ozcelik, B. Cold active pectinase, amylase and protease production by yeast isolates obtained from environmental samples. Extremophiles 2018, 22, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Sabotič, J.; Trcek, T.; Popovic, T.; Brzin, J. Basidiomycetes harbour a hidden treasure of proteolytic diversity. J. Biotechnol. 2007, 128, 297–307. [Google Scholar] [CrossRef]
- Alias, N.; Ahmad Mazian, M.; Salleh, A.B.; Basri, M.; Rahman, R.N.Z.R.A. Molecular cloning and optimization for high level expression of cold-adapted serine protease from antarctic yeast glaciozyma antarctica PI12. Enzyme Res. 2014, 2014, 197938. [Google Scholar] [CrossRef] [Green Version]
- Pi, H.W.; Anandharaj, M.; Kao, Y.Y.; Lin, Y.J.; Chang, J.J.; Li, W.H. Engineering the oleaginous red yeast Rhodotorula glutinis for simultaneous β-carotene and cellulase production. Sci. Rep. 2018, 8, 2–11. [Google Scholar] [CrossRef]
- Mierzejewska, J.; Dąbkowska, K.; Chreptowicz, K.; Sokołowska, A. Hydrolyzed corn stover as a promising feedstock for 2-phenylethanol production by nonconventional yeast. J. Chem. Technol. Biotechnol. 2019, 94, 777–784. [Google Scholar] [CrossRef]
- Baldrian, P.; Valášková, V. Degradation of cellulose by basidiomycetous fungi. FEMS Microbiol. Rev. 2008, 32, 501–521. [Google Scholar] [CrossRef] [Green Version]
- Rong, Y.; Zhang, L.; Chi, Z.; Wang, X. A carboxymethyl cellulase from a marine yeast (Aureobasidium pullulans 98): Its purification, characterization, gene cloning and carboxymethyl cellulose digestion. J. Ocean Univ. China 2015, 14, 913–921. [Google Scholar] [CrossRef]
- Neto, A.A.K.; Borin, G.P.; Goldman, G.H.; de Lima Damásio, A.R.; de Castro Oliveira, J.V. Insights into the plant polysaccharide degradation potential of the xylanolytic yeast Pseudozyma brasiliensis. FEMS Yeast Res. 2016, 16, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnsen, H.R.; Krause, K. Cellulase activity screening using pure carboxymethylcellulose: Application to soluble cellulolytic samples and to plant tissue prints. Int. J. Mol. Sci. 2014, 15, 830–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irazusta, V.; Nieto-Peńalver, C.G.; Cabral, M.E.; Amoroso, M.J.; Figueroa, L.I.C. Relationship among carotenoid production, copper bioremediation and oxidative stress in Rhodotorula mucilaginosa RCL-11. Process Biochem. 2013, 48, 803–809. [Google Scholar] [CrossRef]
- Rivera Velez, S.M. Guide for carotenoid identification in biological samples. J. Nat. Prod. 2016, 79, 1473–1484. [Google Scholar] [CrossRef]
- Hernandez-Almanza, A.; Montanez, J.C.; Aguilar-Gonzalez, M.A.; Martínez-Ávila, C.; Rodríguez-Herrera, R.; Aguilar, C.N. Rhodotorula glutinis as source of pigments and metabolites for food industry. Food Biosci. 2014, 5, 64–72. [Google Scholar] [CrossRef]
- Marova, I.; Szotkowski, M.; Vanek, M.; Rapta, M.; Byrtusova, D.; Mikheichyk, N.; Haronikova, A.; Shapaval, M.C. Utilization of animal fat waste as carbon source by carotenogenic yeasts – a screening study. EuroBiotech J. 2017, 1, 310–318. [Google Scholar] [CrossRef] [Green Version]
- Braunwald, T.; Schwemmlein, L.; Graeff-Hönninger, S.; French, W.T.; Hernandez, R.; Holmes, W.E.; Claupein, W. Effect of different C/N-ratios on carotenoid and lipid production by Rhodotorula glutinis. Appl. Microbiol. Biotechnol. 2013, 6581–6588. [Google Scholar] [CrossRef]
- Sperstad, S.; Lutnæs, B.F.; Stormo, S.K.; Liaaen-Jensen, S.; Bjarne, L. Torularhodin and torulene are the major contributors to the carotenoid pool of marine Rhodosporidium babjevae (Golubev). J. Ind. Microbiol. Biotechnol. 2006, 33, 269. [Google Scholar] [CrossRef]
- Houde, A.; Kademi, A.; Leblanc, D. Lipases and their industrial applications: an overview. Appl. Biochem. Biotechnol. 2004, 118, 155–170. [Google Scholar] [CrossRef]
- Kuhad, R.C.; Gupta, R.; Singh, A. Microbial Cellulases and Their Industrial Applications. Enzyme Res. 2011, 2011, 280696. [Google Scholar] [CrossRef] [Green Version]


| Strain | Source | Identification * | Sequence Length of Genomic DNA (bp) ** / GenBank Accession Number *** |
|---|---|---|---|
| WUT10 | Fermented milk, Antalya, Turkey | Rhodotorula mucilaginosa | 1120 / MN006686 |
| WUT57 | Wild strawberry shrub, Toulouse, France | Rhodotorula graminis | 1111 / MN006688 |
| WUT60 | Pepper, Warsaw, Poland | Rhodotorula mucilaginosa | 1112 / MN006694 |
| WUT61 | Beech tree, Barania Mountain, Poland | Sporobolomyces roseus | 1110 / MN006697 |
| WUT89 | Birch bark, Słowiński National Park, Poland | Cystobasidium laryngis | 1072 / MN006700 |
| WUT92 | Birch bark, Słowiński National Park, Poland | Cystobasidium sp. | 968 / MN006698 |
| WUT103 | Birch bark, Warsaw, Poland | Cystobasidium laryngis | 1103 / MN006701 |
| WUT117 | Mirabelle, Warsaw, Poland | Cystobasidium psychroaquaticum | 1101 / MN006705 |
| WUT128 | Sow-thistle, Turku, Finland | Rhodotorula graminis | 1018 / MN006772 |
| WUT145 | Tree leaf, Cork, Ireland | Cystofilobasidium macerans | 1156 / MN006771 |
| WUT147 | Red grapes, Warsaw, Poland | Rhodotorula graminis | 1005 / MN006773 |
| WUT159 | Apple, Warsaw, Poland | Sporidiobolus pararoseus | 1092 / MN006774 |
| WUT165 | Apple, Warsaw, Poland | Rhodotorula graminis | 1097 / MN006776 |
| WUT167 | Rowanberry, Warsaw, Poland | Rhodotorula mucilaginosa | 1105 / MN006818 |
| WUT182 | Quince, Riga, Latvia | Sporobolomyces roseus | 1095 / MN006819 |
| WUT194 | Grapes, Warsaw, Poland | Rhodotorula graminis | 1107 / MN006820 |
| Strain Number in WUT Collection | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 57 | 60 | 61 | 89 | 92 | 103 | 117 | 128 | 145 | 147 | 159 | 165 | 167 | 182 | 194 | |
| Growth (assimilation) on carbon compounds | ||||||||||||||||
| YNB + GLU | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| YNB + XYL | + | w | w | − | + | + | + | + | + | w | w | − | − | w | − | − |
| YNB + GAL | + | w | + | − | − | + | − | − | + | − | + | w | + | + | − | w |
| YNB + LAC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| YNB + SAC | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + |
| YNB + GLY | + | w | − | + | + | + | + | + | + | w | w | w | − | w | − | w |
| YNB + EtOH | − | − | − | w | + | − | w | w | + | w | w | − | − | w | − | w |
| YNB + CEL | − | + | − | w | w | + | + | + | + | + | + | + | + | + | w | w |
| YNB + MAL | + | + | + | + | − | − | − | − | + | + | + | + | + | + | + | − |
| Range of temperature growth (YPD medium, 2–3 days) | ||||||||||||||||
| [°C] | 4–30 | 4–30 | 4–30 | 4–25 | 4–25 | 4–25 | 4–25 | 4–25 | 4–30 | 4–25 | 4–30 | 4–30 | 4–25 | 4–37 | 4–30 | 4–30 |
| Extracellular enzymes production | ||||||||||||||||
| Lipase activity a | + | + | + | + | w | w | w | w | + | + | + | − | + | + | w | + |
| Protease activity b | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − |
| Cellulase activity c | − | − | − | + | − | − | − | − | − | + | − | + | − | − | + | − |
| Strain | Halo Diameter [mm] | CMC Activity* [µmol/mg Protein*min] |
|---|---|---|
| WUT61 | 9.7 ± 0.5 | 0.65 ± 0.21 |
| WUT145 | 8.7 ± 1.4 | 66.23 ± 0.15 |
| WUT159 | 11.0 ± 0.0 | 0.30 ± 0.10 |
| WUT182 | 10.0 ± 1.4 | 3.54 ± 0.76 |
| WUT73 (K+) | 11.3 ± 0.9 | 1.55 ± 0.09 |
| Strain | X [g/L] | Residual Glucose [g/L] | L [mg] | CC [mg/g Biomass] | YC [mg/L] | Detected Carotenoids * |
|---|---|---|---|---|---|---|
| First test group (at 25 °C) | ||||||
| WUT10 | 6.86 ± 0.24 | nd | 1.85 ± 0.76 | 0.23 ± 0.02 | 1.49 ± 0.10 | torularhodin |
| WUT57 | 4.55 ± 0.22 | nd | a | 0.25 ± 0.02 | 1.10 ± 0.10 | torularhodin, β-carotene, γ-carotene |
| WUT60 | 6.91 ± 0.10 | nd | 8.95 ± 0.99 | 0.11 ± 0.01 | 0.66 ± 0.10 | torularhodin |
| WUT61 | 7.37 ± 0.21 | nd | a | 0.16 ± 0.00 | 1.18 ± 0.00 | torularhodin, β-carotene, γ-carotene, torulene |
| WUT89 | 4.65 ± 0.29 | 9.09 ± 0.15 | 8.30 ± 0.42 | 0.09 ± 0.01 | 0.41 ± 0.09 | β-carotene, γ-carotene, torulene, torularhodin |
| WUT92 | 7.55 ± 0.25 | nd | 22.30 ± 1.56 | 0.32 ± 0.04 | 2.62 ± 0.03 | torularhodin, torulene, β-carotene, γ-carotene |
| WUT103 | 4.51 ± 0.17 | 9.25 ± 0.67 | 5.10 ± 0.72 | 0.05 ± 0.01 | 0.25 ± 0.05 | β-carotene, γ-carotene, torulene, torularhodin |
| WUT117 | 4.42 ± 0.06 | 6.50 ± 0.51 | 8.00 ± 0.57 | 0.06 ± 0.01 | 0.31 ± 0.04 | β-carotene, γ-carotene, torularhodin, torulene |
| Second test group (at 22 °C) | ||||||
| WUT128 | 7.10 ± 0.26 | nd | 25.70 ± 5.52 | 0.93 ± 0.10 | 6.61 ± 0.92 # | torularhodin, β-carotene, γ-carotene, torulene |
| WUT145 | 6.70 ± 0.90 | nd | 7.40 ± 0.42 | 0.06 ± 0.012 | 0.62 ± 0.02 | torulene |
| WUT147 | 9.70 ± 1.27 | nd | 13.85 ± 3.61 | 0.60 ± 0.06 | 6.00 ± 0.52 | torularhodin, β-carotene, γ-carotene, torulene |
| WUT159 | 7.17 ± 0.23 | nd | 13.45 ± 3.89 | 0.75 ± 0.06 | 5.35 ± 0.51 | torularhodin, β-carotene, γ-carotene, torulene |
| WUT165 | 9.07 ± 0.75 | nd | 4.20 ± 0.28 | 0.35 ± 0.05 | 3.06 ± 0.09 | torularhodin, β-carotene, torulene |
| WUT167 | 8.67 ± 0.32 | nd | 8.95 ± 1.34 | 0.34 ± 0.03 | 3.09 ± 0.21 | torularhodin, β-carotene, torulene |
| WUT182 | 9.23 ± 0.21 | nd | 19.70 ± 0.28 | 1.12 ± 0.01 | 10.33 ± 0.24 # | torulene, torularhodin, β-carotene, γ-carotene |
| WUT194 | 5.97 ± 0.15 | nd | 7.70 ± 0.99 | 0.35 ± 0.05 | 3.06 ± 0.09 | torularhodin, β-carotene, γ-carotene |
| Strain | X [g/L] | Residual Glucose [g/L] | L [mg] | CC [mg/g Biomass] | YC [mg/L] | Detected Carotenoids |
|---|---|---|---|---|---|---|
| 15 °C | ||||||
| WUT89 | 7.58 ± 0.28 | 7.45 ± 0.24 | 34.00 ± 6.82 | 0.10 ± 0.03 | 0.73 ± 0.22 # | γ-carotene |
| WUT92 | 6.48 ± 0.38 | 9.95 ± 0.18 | 12.35 ± 1.71 | 0.14 ± 0.00 | 0.92 ± 0.06 | γ-carotene |
| WUT117 | 5.13 ± 0.32 | 7.42 ± 0.13 | 45.40 ± 3.48 | 0.54 ± 0.08 | 3.05 ± 0.45 # | γ-carotene |
| 20 °C | ||||||
| WUT89 | 7.15 ± 0.82 | 4.83 ± 0.24 | 137.70 ± 12.59 | 0.10 ± 0.02 | 0.58 ± 0.14 | γ-carotene |
| WUT92 | 6.93 ± 0.42 | 6.85 ± 0.27 | 87.50 ± 5.82 | 0.15 ± 0.04 | 0.24 ± 0.02 | γ-carotene, torularhodin |
| WUT117 | 6.43 ± 0.30 | 3.78 ± 0.07 | 85.35 ± 4.19 | 0.14 ± 0.09 | 1.05 ± 0.30 | γ-carotene |
| Strain | X [g/L] | CC [mg/g Biomass] | YC [mg/L] | Carotenoid Distribution [mg/L] | |||
|---|---|---|---|---|---|---|---|
| β-Carotene | γ-Carotene | Torulene | Torularhodin | ||||
| 30 g/L glucose | |||||||
| 10 | 10.75 ± 0.35 # | 0.04 ± 0.00 | 0.38 ± 0.05 | 0.06 ± 0.01 | 0 | 0 | 0.32 ± 0.04 |
| 60 | 10.43 ± 0.55 | 0.02 ± 0.00 | 0.17 ± 0.03 | 0 | 0.04 ± 0.01 | 0 | 0.15 ± 0.02 |
| 145 | 10.43 ± 1.27 | 0.08 ± 0.03 | 0.95 ± 0.06 | 0 | 0.33 ± 0.02 | 0.24 ± 0.04 | 0.13 ± 0.01 |
| 40 g/L glucose | |||||||
| 10 | 16.60 ± 0.70 | 0.04 ± 0.01 | 0.64 ± 0.11 | 0 | 0.07 ± 0.01 | 0 | 0.64 ± 0.04 |
| 60 | 13.37 ± 0.55 | 0.04 ± 0.01 | 0.39 ± 0.06 | 0 | 0.05 ± 0.01 | 0 | 0.34 ± 0.05 |
| 145 | 13.37 ± 0.55 | 0.13 ± 0.01 | 1.72 ± 0.08 # | 0 | 0.51 ± 0.02 | 0.56 ± 0.04 | 0.23 ± 0.03 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chreptowicz, K.; Mierzejewska, J.; Tkáčová, J.; Młynek, M.; Čertik, M. Carotenoid-Producing Yeasts: Identification and Characteristics of Environmental Isolates with a Valuable Extracellular Enzymatic Activity. Microorganisms 2019, 7, 653. https://doi.org/10.3390/microorganisms7120653
Chreptowicz K, Mierzejewska J, Tkáčová J, Młynek M, Čertik M. Carotenoid-Producing Yeasts: Identification and Characteristics of Environmental Isolates with a Valuable Extracellular Enzymatic Activity. Microorganisms. 2019; 7(12):653. https://doi.org/10.3390/microorganisms7120653
Chicago/Turabian StyleChreptowicz, Karolina, Jolanta Mierzejewska, Jana Tkáčová, Mateusz Młynek, and Milan Čertik. 2019. "Carotenoid-Producing Yeasts: Identification and Characteristics of Environmental Isolates with a Valuable Extracellular Enzymatic Activity" Microorganisms 7, no. 12: 653. https://doi.org/10.3390/microorganisms7120653
APA StyleChreptowicz, K., Mierzejewska, J., Tkáčová, J., Młynek, M., & Čertik, M. (2019). Carotenoid-Producing Yeasts: Identification and Characteristics of Environmental Isolates with a Valuable Extracellular Enzymatic Activity. Microorganisms, 7(12), 653. https://doi.org/10.3390/microorganisms7120653