Presence of Mycobacterium avium Subspecies paratuberculosis Monitored Over Varying Temporal and Spatial Scales in River Catchments: Persistent Routes for Human Exposure
Abstract
:1. Introduction
- to determine whether the overall distribution and presence of Map in the catchment has changed over time when farming practices and management of stock remain the same,
- to test whether waste water treatment effluent is a significant input of Map in the river,
- to assess whether presently used sampling regimes to detect Map in the environment underestimate its prevalence,
- to quantify the presence of Map in rivers, and
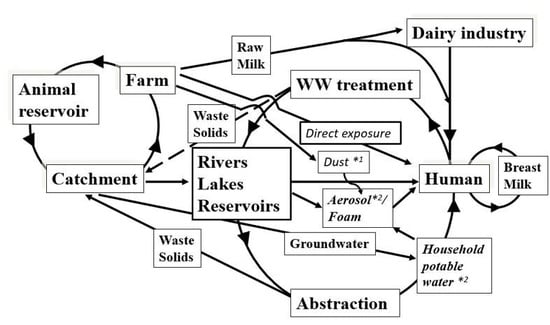
- to extend our catchment model [41].
2. Materials and Methods
2.1. Study Areas
2.1.1. River Tywi, Wales, UK
2.1.2. Eden Catchment, Cumbria, UK
2.1.3. Sampling Sites
2.2. Sample Processing
2.3. Detection of Map by PCR
2.4. Statistical Analysis of Map and the Tywi and Eden Catchments
3. Results and Discussion
3.1. Overview
3.2. Temporal and Spatial Monitoring and Quantification of Map in Rivers
3.2.1. Weekly Monitoring of Map in the River Tywi Catchment
3.2.2. The Contribution of Waste Water Treatment Works (WWTWs) to the Presence of Map in Rivers
3.2.3. Increased Resolution ‘Fine’ Sampling Over a Short Time Period
3.2.4. Sampling Period and Map Presence
3.3. Quantification of Map
3.4. Overview Map in Welsh River Catchments
3.5. Sampling Map in New Locations
3.5.1. Estuarine Samples
3.5.2. Eden Catchment Field Beck Samples
3.5.3. Natural River Foam
4. Conclusions
- the overall distribution and presence of Map in the catchment has not changed over a 10-year period and that associated farming practices and management of stock have remained the same;
- that the effluent small waste water treatment works does not significantly input of Map into the river, but large city-based waste treatment works do have a significant input;
- using a variety of temporal and spatial sampling regimes from monthly single sample points to fine sampling every 30 min, that weekly monitoring from a single carefully chosen location, adequately describes the presence of Map in rivers emerging from defined catchments or sub-catchments. Monitoring due to regulatory policy could become mandatory if Map is confirmed as a human pathogen and we suggest a monitoring scheme that is feasible;
- for the first time, Map concentration in rivers has been assessed and numbers ranged up to 108 cell equivalents L−1;
Author Contributions
Funding
Conflicts of Interest
References
- Thorel, M.F.; Krichevsky, M.; Levyfrebault, V.V. Numerical taxonomy of mycobactin-dependent mycobacteria, emended description of mycobacterium avium, and description of mycobacterium avium subsp avium subsp-nov, mycobacterium avium subsp paratuberculosis subsp-nov, and mycobacterium avium subsp silvaticum subsp-nov. Int. J. Syst. Bact. 1990, 40, 254–260. [Google Scholar]
- Chacon, O.; Bermudez, L.E.; Barletta, R.G. Johne’s disease, inflammatory bowel disease, and mycobacterium paratuberculosis. Annu. Rev. Microbiol. 2004, 58, 329–363. [Google Scholar] [CrossRef]
- Over, K.; Crandall, P.G.; O’Bryan, C.A.; Ricke, S.C. Current perspectives on mycobacterium avium subsp. Paratuberculosis, johne’s disease, and crohn’s disease: A review. Crit. Rev. Microbiol. 2011, 37, 141–156. [Google Scholar] [PubMed]
- Arru, G.; Caggiu, E.; Paulus, K.; Sechi, G.P.; Mameli, G.; Sechi, L.A. Is there a role for mycobacterium avium subspecies paratuberculosis in parkinson’s disease? J. Neuroimmunol. 2016, 293, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Bo, M.; Erre, G.L.; Niegowska, M.; Piras, M.; Taras, L.; Longu, M.G.; Passiu, G.; Sechi, L.A. Interferon regulatory factor 5 is a potential target of autoimmune response triggered by epstein-barr virus and mycobacterium avium subsp. Paratuberculosis in rheumatoid arthritis: Investigating a mechanism of molecular mimicry. Clin. Exp. Rheumatol. 2018, 36, 376–381. [Google Scholar]
- Bo, M.; Niegowska, M.; Arru, G.; Sechi, E.; Mariotto, S.; Mancinelli, C.; Farinazzo, A.; Alberti, D.; Gajofatto, A.; Ferrari, S.; et al. Mycobacterium avium subspecies paratuberculosis and myelin basic protein specific epitopes are highly recognized by sera from patients with neuromyelitis optica spectrum disorder. J. Neuroimmunol. 2018, 318, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Niegowska, M.; Wajda-Cuszlag, M.; Stępień-Ptak, G.; Trojanek, J.; Michałkiewicz, J.; Szalecki, M.; Sechi, L.A. Anti-herv-wenv antibodies are correlated with seroreactivity against mycobacterium avium subsp. Paratuberculosis in children and youths at t1d risk. Sci. Rep. 2019, 9, 6282. [Google Scholar]
- Mameli, G.; Cocco, E.; Frau, J.; Arru, G.; Caggiu, E.; Marrosu, M.G.; Sechi, L.A. Serum baff levels, methypredsinolone therapy, epstein-barr virus and mycobacterium avium subsp paratuberculosis infection in multiple sclerosis patients. Sci. Rep. 2016, 6, 29268. [Google Scholar] [CrossRef]
- Niegowska, M.; Delitala, A.; Pes, G.M.; Delitala, G.; Sechi, L.A. Increased seroreactivity to proinsulin and homologous mycobacterial peptides in latent autoimmune diabetes in adults. PLoS ONE 2017, 12, e0176584. [Google Scholar] [CrossRef] [PubMed]
- Slavin, Y.N.; Bo, M.; Caggiu, E.; Sechi, G.; Arru, G.; Bach, H.; Sechi, L.A. High levels of antibodies against ptpa and pkng secreted by mycobacterium avium ssp paratuberculosis are present in neuromyelitis optica spectrum disorder and multiple sclerosis patients. J. Neuroimmunol. 2018, 323, 49–52. [Google Scholar] [CrossRef]
- Zamani, S.; Zali, M.R.; Aghdaei, H.A.; Sechi, L.A.; Niegowska, M.; Caggiu, E.; Keshavarz, R.; Mosavari, N.; Feizabadi, M.M. Mycobacterium avium subsp paratuberculosis and associated risk factors for inflammatory bowel disease in iranian patients. Gut Pathog. 2017, 9, 1. [Google Scholar] [CrossRef] [PubMed]
- Kuenstner, J.T.; Naser, S.; Chamberlin, W.; Borody, T.; Graham, D.Y.; McNees, A.; Hermon-Taylor, J.; Hermon-Taylor, A.; Dow, C.T.; Thayer, W.; et al. The consensus from the mycobacterium avium ssp. Paratuberculosis (map) conference 2017. Front. Public Health 2017, 5, 208. [Google Scholar] [PubMed]
- Buergelt, C.D.; Hall, C.; McEntee, K.; Duncan, J.R. Pathological evaluation of para-tuberculosis in naturally infected cattle. Vet. Path. 1978, 15, 196–207. [Google Scholar] [CrossRef]
- Nielsen, S.S.; Toft, N. A review of prevalences of paratuberculosis in farmed animals in europe. Prev. Vet. Med. 2009, 88, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Rathnaiah, G.; Zinniel, D.K.; Bannantine, J.P.; Stabel, J.R.; Grohn, Y.T.; Collins, M.T.; Barletta, R.G. Pathogenesis, molecular genetics, and genomics of mycobacterium avium subsp. Paratuberculosis, the etiologic agent of johne’s disease. Front. Vet. Sci. 2017, 4, 187. [Google Scholar] [CrossRef] [PubMed]
- Feller, M.; Huwiler, K.; Stephan, R.; Altpeter, E.; Shang, A.; Furrer, H.; Pfyffer, G.E.; Jemmi, T.; Baumgartner, A.; Egger, M. Mycobacterium avium subspecies paratuberculosis and crohn’s disease: A systematic review and meta-analysis. Lancet Infect. Dis. 2007, 7, 607–613. [Google Scholar] [CrossRef]
- Chiodini, R.J.; Chamberlin, W.M.; Sarosiek, J.; McCallum, R.W. Crohn’s disease and the mycobacterioses: A quarter century later. Causation or simple association? Crit. Rev. Microbiol. 2012, 38, 52–93. [Google Scholar] [CrossRef]
- Timms, V.J.; Daskalopoulos, G.; Mitchell, H.M.; Neilan, B.A. The association of mycobacterium avium subsp paratuberculosis with inflammatory bowel disease. PLoS ONE 2016, 11, e0148731. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.A.; Pilli, S.; Surendranath, A.; Rampal, R.; Chauhan, S.K.; Tiwari, V.; Mouli, V.P.; Kedia, S.; Nayak, B.; Das, P.; et al. Prevalence and association of mycobacterium avium subspecies paratuberculosis with disease course in patients with ulcero-constrictive ileocolonic disease. PLoS ONE 2016, 11, e0152063. [Google Scholar] [CrossRef]
- Singh, S.V.; Kuenstner, J.T.; Davis, W.C.; Agarwal, P.; Kumar, N.; Singh, D.; Gupta, S.; Chaubey, K.K.; Kumar, A.; Misri, J.; et al. Concurrent resolution of chronic diarrhea likely due to crohn’s disease and infection with mycobacterium avium paratuberculosis. Front. Med. 2016, 3, 49. [Google Scholar] [CrossRef]
- Waddell, L.A.; Rajic, A.; Stark, K.D.C.; McEwen, S.A. The potential public health impact of mycobacterium avium ssp. Paratuberculosis: Global opinion survey of topic specialists. Zoonoses Public Health 2016, 63, 212–222. [Google Scholar] [CrossRef]
- Bull, T.J.; McMinn, E.J.; Sidi-Boumedine, K.; Skull, A.; Durkin, D.; Neild, P.; Rhodes, G.; Pickup, R.; Hermon-Taylor, J. Detection and verification of mycobacterium avium subsp. Paratuberculosis in fresh ileocolonic mucosal biopsy specimens from individuals with and without crohn’s disease. J. Clin. Microbiol. 2003, 41, 2915–2923. [Google Scholar]
- Naser, S.A.; Ghobrial, G.; Romero, C.; Valentine, J.F. Culture of mycobacterium avium subsp. Paratuberculosis from the blood of patients with crohn’s disease. Lancet 2004, 364, 1039–1044. [Google Scholar]
- Naser, S.A.; Schwartz, D.; Shafran, I. Isolation of mycobacterium avium subsp paratuberculosis from breast milk of crohn’s disease patients. Am. J. Gastroenterol. 2000, 95, 1094–1095. [Google Scholar] [CrossRef]
- Norgard, B.M.; Nielsen, J.; Fonager, K.; Kjeldsen, J.; Jacobsen, B.A.; Qvist, N. The incidence of ulcerative colitis (1995-2011) and crohn’s disease (1995-2012)—Based on nationwide danish registry data. J. Crohns Colitis 2014, 8, 1274–1280. [Google Scholar] [CrossRef]
- Studd, C.; Cameron, G.; Beswick, L.; Knight, R.; Hair, C.; McNeil, J.; Desmond, P.; Wilson, J.; Connell, W.; Bell, S. Never underestimate inflammatory bowel disease: High prevalence rates and confirmation of high incidence rates in australia. J. Gastroenterol. Hepatol. 2016, 31, 81–86. [Google Scholar] [CrossRef]
- Wilson, J.; Hair, C.; Knight, R.; Catto-Smith, A.; Bell, S.; Kamm, M.; Desmond, P.; McNeil, J.; Connell, W. High incidence of inflammatory bowel disease in australia: A prospective population-based australian incidence study. Inflamm. Bowel Dis. 2010, 16, 1550–1556. [Google Scholar] [CrossRef]
- Galeone, C.; Pelucchi, C.; Barbera, G.; Citterio, C.; La Vecchia, C.; Franchi, A. Crohn’s disease in italy: A critical review of the literature using different data sources. Dig. Liver Dis. 2017, 49, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Floyd, D.N.; Langham, S.; Severac, H.C.; Levesque, B.G. The economic and quality-of-life burden of crohn’s disease in europe and the united states, 2000 to 2013: A systematic review. Dig. Dis. Sci. 2015, 60, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, J.; Sykora, J.; Cvalinova, D.; Pomahacova, R.; Kleckova, J.; Kryl, M.; Vcelak, P. Inflammatory bowel disease incidence in czech children: A regional prospective study, 2000-2015. World J. Gastroenterol. 2017, 23, 4090–4101. [Google Scholar] [CrossRef]
- Lehtinen, P.; Ashorn, M.; Iltanen, S.; Jauhola, R.; Jauhonen, P.; Kolho, K.L.; Auvinen, A. Incidence trends of pediatric inflammatory bowel disease in finland, 1987-2003, a nationwide study. Inflamm. Bowel Dis. 2011, 17, 1778–1783. [Google Scholar] [CrossRef]
- Pozler, O.; Maly, J.; Bonova, O.; Dedek, P.; Fruhauf, P.; Havlickova, A.; Janatova, T.; Jimramovsky, F.; Klimova, L.; Klusacek, D.; et al. Incidence of crohn disease in the czech republic in the years 1990 to 2001 and assessment of pediatric population with inflammatory bowel disease. J. Pediatr. Gastroenterol. Nutr. 2006, 42, 186–189. [Google Scholar] [CrossRef]
- Olsen, I.; Siguroardottir, O.G.; Djonne, B. Paratuberculosis with special reference to cattle—A review. Vet. Q. 2002, 24, 12–28. [Google Scholar] [CrossRef]
- Sorensen, O.; Rawluk, S.; Wu, J.; Manninen, K.; Ollis, G. Mycobacterium paratuberculosis in dairy herds in alberta. Can. Vet. J. 2003, 44, 221–226. [Google Scholar]
- United States Department of Agriculture. Johne’s Disease on U. S. Dairies. Available online: http://www.aphis.usda.gov/animal_health/nahms/dairy/downloads/dairy07/Dairy07_is_Johnes.pdf (accessed on 9 May 2019).
- Caldow, G.; Strain, S.A.J.; Chapman, Z.; Kemp, R.; Cook, A.J. A survey to estimate the herd level prevalence of paratuberculosis in the dairy herd of the united kingdom. Cattle Pract. 2007, 15, 169–171. [Google Scholar]
- Kemp, R.; Caldow, G.; Strain, S. Herd prevalence of johne’s disease. Vet. Rec. 2006, 159, 572. [Google Scholar] [CrossRef] [PubMed]
- Velasova, M.; Damaso, A.; Prakashbabu, B.C.; Gibbons, J.; Wheelhouse, N.; Longbottom, D.; Van Winden, S.; Green, M.; Guitian, J. Herd-level prevalence of selected endemic infectious diseases of dairy cows in great britain. J. Dairy Sci. 2017, 100, 9215–9233. [Google Scholar] [CrossRef]
- DEFRA, Department of the Environment Food and Rural Affairs (UK). An Integrated Strategy to Determine the Herd Level Prevalence of Johne’s Disease in the UK Dairy Herd. Available online: http://archive.defra.gov.uk/foodfarm/farmanimal/diseases/atoz/documents/johnes-report0911.pdf (accessed on 9 May 2019).
- Clarke, C.J. The pathology and pathogenesis of paratuberculosis in ruminants and other species. J. Comp. Pathol. 1997, 116, 217–261. [Google Scholar] [CrossRef]
- Pickup, R.W.; Rhodes, G.; Bull, T.J.; Arnott, S.; Sidi-Boumedine, K.; Hurley, M.; Hermon-Taylor, J. Mycobacterium avium subsp. Paratuberculosis in lake catchments, in river water abstracted for domestic use, and in effluent from domestic sewage treatment works: Diverse opportunities for environmental cycling and human exposure. Appl. Environ. Microbiol. 2006, 72, 4067–4077. [Google Scholar] [CrossRef]
- Whittington, R.J.; Marsh, I.B.; Reddacliff, L.A. Survival of mycobacterium avium subsp paratuberculosis in dam water and sediment. Appl. Environ. Microbiol. 2005, 71, 5304–5308. [Google Scholar] [CrossRef]
- Pickup, R.W.; Rhodes, G.; Sidi-Boumedine, K.; Arnott, S.; Bull, T.; Weightman, A.J.; Hermon-Taylor, J. Mycobacterium avium subspecies paratuberculosis in the catchment and water of the river taff in South Wales, UK and its potential relationship to clustering of crohn’s disease in the city of cardiff. Appl. Environ. Microbiol. 2005, 71, 2130–2139. [Google Scholar] [CrossRef]
- Rhodes, G.; Richardson, H.; Hermon-Taylor, J.; Weightman, A.; Higham, A.; Pickup, R. Mycobacterium avium subspecies paratuberculosis: Human exposure through environmental and domestic aerosols. Pathogens 2014, 3, 577–595. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, G.; Henrys, P.; Thomson, B.C.; Pickup, R.W. Mycobacterium avium subspecies paratuberculosis is widely distributed in british soils and waters: Implications for animal and human health. Envir. Microbiol. 2013, 15, 2761–2774. [Google Scholar]
- Owen, G.J.; Perks, M.T.; Benskin, C.M.H.; Wilkinson, M.E.; Jonczyk, J.; Quinn, P.F. Monitoring agricultural diffuse pollution through a dense monitoring network in the river eden demonstration test catchment, cumbria, UK. Area 2012, 44, 443–453. [Google Scholar] [CrossRef]
- Schilling, K.; Zessner, M. Foam in the aquatic environment. Water Res. 2011, 45, 4355–4366. [Google Scholar] [CrossRef] [PubMed]
- Schoenenbrucher, H.; Abdurnawjood, A.; Failing, K.; Buelte, M. New triplex real-time pcr assay for detection of mycobacterium avium subsp paratuberculosis in bovine feces. Appl. Environ. Microbiol. 2008, 74, 2751–2758. [Google Scholar] [CrossRef] [PubMed]
- Slana, I.; Kralik, P.; Kralova, A.; Pavlik, I. On-farm spread of mycobacterium avium subsp paratuberculosis in raw milk studied by is900 and f57 competitive real time quantitative pcr and culture examination. Int. J. Food Microbiol. 2008, 128, 250–257. [Google Scholar] [CrossRef]
- Rhodes, G.; Fluri, A.; Gerber, M.; Henderson, A.; Ruefenacht, A.; Pickup, R.W. Detection of mycobacterium immunogenum by real-time quantitative taqman pcr. J. Microbiol. Meth. 2008, 73, 266–268. [Google Scholar] [CrossRef]
- Poupart, P.; Coene, M.; Vanheuverswyn, H.; Cocito, C. Preparation of a specific rna probe for detection of mycobacterium paratuberculosis and diagnosis of johnes disease. J. Clin. Microbiol. 1993, 31, 1601–1605. [Google Scholar]
- Li, L.L.; Bannantine, J.P.; Zhang, Q.; Amonsin, A.; May, B.J.; Alt, D.; Banerji, N.; Kanjilal, S.; Kapur, V. The complete genome sequence of mycobacterium avium subspecies paratuberculosis. Proc. Natn. Acad. Sci. USA 2005, 102, 12344–12349. [Google Scholar] [CrossRef] [PubMed]
- McCullagh, P.; Nelder, J.A. Generalized Linear Models, 2nd ed.; Monographs on Statistics and Applied Probability 37; Chapman and Hall/CRC: Boca Raton, FL, USA, 1989. [Google Scholar]
- Team, R.D.C. R: A Language and Environment for Statistical Computing. Available online: http://www.R-project.org (accessed on 9 May 2019).
- Mardia, K.V.; Kent, J.T.; Bibby, J.M. Multivariate Analysis; Academic Press: Cambridge, MA, USA, 1979. [Google Scholar]
- Herthnek, D.; Englund, S.; Willemsen, P.T.J.; Bolske, G. Sensitive detection of myobacterium avium subsp paratuberculosis in bovine semen by real-time pcr. J. Appl. Microbiol. 2006, 100, 1095–1102. [Google Scholar] [CrossRef] [PubMed]
- Pavlik, I.; Horvathova, A.; Dvorska, L.; Bartl, J.; Svastova, P.; du Maine, R.; Rychlik, I. Standardisation of restriction fragment length polymorphism analysis for mycobacterium avium subspecies paratuberculosis. J. Microbiol. Meth. 1999, 38, 155–167. [Google Scholar] [CrossRef]
- Terry, J.A.; Benskin, C.M.H.; Eastoe, E.F.; Haygarth, P.M. Temporal dynamics between cattle in-stream presence and suspended solids in a headwater catchment. Environ. Sci. Process. Impacts 2014, 16, 1570–1577. [Google Scholar] [CrossRef]
- Toth, J.D.; Aceto, H.W.; Rankin, S.C.; Dou, Z. Short communication: Survey of animal-borne pathogens in the farm environment of 13 dairy operations. J. Dairy Sci. 2013, 96, 5756–5761. [Google Scholar] [CrossRef]
- Petrovski, S.; Dyson, Z.; Quill, E.; SJ, M.; Daniel Tillett, D.; Seviour, R. An examination of the mechanisms for stable foam formation in activated sludge systems. Water Res. 2011, 45, 2146–2154. [Google Scholar] [CrossRef]
- Napolitano, G.E.; Richmond, J.E. Enrichment of biogenic lipids, hydrocarbons and pcbs in stream-surface foams. Environ. Toxicol. Chem. 1995, 14, 197–201. [Google Scholar] [CrossRef]
- Maynard, N.G. Aquatic foams as an ecological habitat. Z. Allg. Mikrobiol. 1968, 8, 119–127. [Google Scholar] [CrossRef]
- Wolf, R.; Donate, K.; Khol, J.L.; Barkema, H.W.; Kastelic, J.; Wagner, P. Detection of mycobacterium avium subspecies paratuberculosis infected cattle herds using environmental samples: A review. Berl. Munch. Tierarztl. Wochenschr. 2017, 130, 4–12. [Google Scholar]
- Thirunavukkarasu, S.; Plain, K.M.; de Silva, K.; Marais, B.J.; Whittington, R.J. Applying the one health concept to mycobacterial research overcoming parochialism. Zoonoses Public Health 2017, 64, 401–422. [Google Scholar] [CrossRef] [PubMed]
- Sechi, L.A.; Dow, C.T. Mycobacterium avium ss. Paratuberculosis—Zoonosis the hundred year war—Beyond crohn’s disease. Front. Immunol. 2015, 6, 1–8. [Google Scholar] [CrossRef]




| Sample Site | UK OSGR | Sample Type | Date | Samples n = |
|---|---|---|---|---|
| Tywi upstream of WWTW * (A) | SN616212 | River water | Monthly | 15 |
| Tywi downstream of WWTW * (B) | SN614211 | River water | Monthly | 15 |
| Tywi upstream (5) | SN533212 | River water | Weekly | 65 |
| Cothi (6) | SN505217 | River water | Weekly | 61 |
| Cothi upstream of WWTW * (C) | SN506214 | River water | Monthly | 14 |
| Cothi downstream of WWTW * (D) | SN502210 | River water | Monthly | 14 |
| Tywi downstream (Nantgaredig; 7) | SN491203 | River water | Weekly | 64 |
| River | Height (m) | Flow (m3 s−1) | Rainfall (Days before Sampling) | ||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 6 | 9 | |||
| Cothi | 0.0026 | 0.0019 | 0.003 | ns | 0.01 | ns | 0.018 |
| Tywi upstream | 0.00013 | 0.00008 | 0.02 | 0.000006 | 0.01 | 0.034 | ns |
| Tywi downstream | 0.037 | 0.029 | 0.03 | 0.00002 | ns | 0.011 | ns |
| Sample Site | OSGR | Sample Type | Sampling Date |
|---|---|---|---|
| Ford (1) | SN595222 | Foam | 24.07.2012 |
| Field drain water | 23.04.2013 | ||
| River water | 24.07/20/11.2012 | ||
| River water | 23.04.2013 | ||
| Dulus pre-tributary (2) | SN550212 | River water | 24.07/20.11.2012 |
| River water | 23.04.2013 | ||
| Dulas side tributary (3) | SN550212 | River water | 24.07/20.11.2012 |
| River water | 23.04.2013 | ||
| Dulas downstream (4) | SN550212 | River water | 24.07/20.11.2012 |
| River water | 23.04.2013 | ||
| Sediment | 23.04.2013 | ||
| Foam | 24.07.2012 | ||
| Tywi upstream (5) | SN533212 | Sediment | 24.07.2012 |
| Sediment | 23.04.2013 | ||
| River water (FS) | 24.07/20.11.2012 | ||
| River water (FS) | 23.04.2013 | ||
| Cothi (6) | SN505217 | River water | 24.07/20.11.2012 |
| River water | 23.04.2013 | ||
| Nantgaredig bridge (7) Tywi downsteam | SN494204 | River water River water | 24.07/20.11.2012 23.04.2013 |
| Sub-Catchments | Sample Site | OSRG |
|---|---|---|
| Morland | Long Sike | NY581196 |
| Sleagill Beck | NY596190 | |
| Newby Beck | NY597212 | |
| Darce | Lowthwaite Beck | NY409236 |
| Thackthwaite Beck at Nabend | NY411253 | |
| Mell Fell Beck | NY407244 | |
| Pow | Pow Beck at Beckhouse Bridge | NY422469 |
| Unnamed tributary Pow Beck | NY386500 | |
| Pow Beck at Green Lane | NY386500 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Richardson, H.; Rhodes, G.; Henrys, P.; Sedda, L.; Weightman, A.J.; Pickup, R.W. Presence of Mycobacterium avium Subspecies paratuberculosis Monitored Over Varying Temporal and Spatial Scales in River Catchments: Persistent Routes for Human Exposure. Microorganisms 2019, 7, 136. https://doi.org/10.3390/microorganisms7050136
Richardson H, Rhodes G, Henrys P, Sedda L, Weightman AJ, Pickup RW. Presence of Mycobacterium avium Subspecies paratuberculosis Monitored Over Varying Temporal and Spatial Scales in River Catchments: Persistent Routes for Human Exposure. Microorganisms. 2019; 7(5):136. https://doi.org/10.3390/microorganisms7050136
Chicago/Turabian StyleRichardson, Hollian, Glenn Rhodes, Peter Henrys, Luigi Sedda, Andrew J. Weightman, and Roger W. Pickup. 2019. "Presence of Mycobacterium avium Subspecies paratuberculosis Monitored Over Varying Temporal and Spatial Scales in River Catchments: Persistent Routes for Human Exposure" Microorganisms 7, no. 5: 136. https://doi.org/10.3390/microorganisms7050136
APA StyleRichardson, H., Rhodes, G., Henrys, P., Sedda, L., Weightman, A. J., & Pickup, R. W. (2019). Presence of Mycobacterium avium Subspecies paratuberculosis Monitored Over Varying Temporal and Spatial Scales in River Catchments: Persistent Routes for Human Exposure. Microorganisms, 7(5), 136. https://doi.org/10.3390/microorganisms7050136