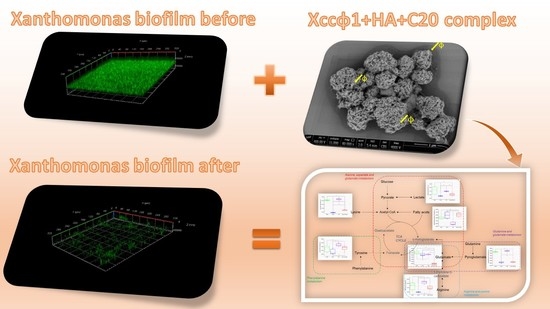
Bacteriophages Promote Metabolic Changes in Bacteria Biofilm
Abstract
:1. Introduction
2. Material and Methods
2.1. Isolation and Growth of Xcc Phages
2.2. Eicosanoic Acid Activity against Biofilm
2.3. Preparation of Supernatants for Metabolic Analysis
2.4. NMR Spectroscopy
2.5. Multivariate Data Analysis
2.6. Pathway Analysis
3. Results
3.1. Phage Xccφ1, Hydroxyapatite, and Eicosanoic Acid Modulate Xcc Biofilm
3.2. NMR Analysis: Class Discrimination
3.3. Pathway Analysis
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- European Antimicrobial Resistance Surveillance Network (EARS-Net). Available online: https://www.ecdc.europa.eu/en/about-us/partnerships-and-networks/disease-and-laboratory-networks/ears-net (accessed on 27 March 2020).
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [Green Version]
- Stewart, P.S.; Costerton, J.W. Antibiotic resistance of bacteria in biofilms. Lancet 2001, 358, 135–138. [Google Scholar] [CrossRef]
- Burmølle, M.; Thomsen, T.R.; Fazli, M.; Dige, I.; Christensen, L.; Homøe, P.; Tvede, M.; Nyvad, B.; Tolker-Nielsen, T.; Givskov, M.; et al. Biofilms in chronic infections—A matter of opportunity—Monospecies biofilms in multispecies infections. FEMS Immunol. Med. Microbiol. 2010, 59, 324–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boudjemaa, R.; Cabriel, C.; Dubois-Brissonnet, F.; Bourg, N.; Dupuis, G.; Gruss, A.; Lévêque-Fort, S.; Briandet, R.; Fontaine-Aupart, M.-P.; Steenkeste, K. Impact of bacterial membrane fatty acid composition on the failure of daptomycin to kill staphylococcus aureus. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [Green Version]
- Craft, K.M.; Nguyen, J.M.; Berg, L.J.; Townsend, S.D. Methicillin-resistant: Staphylococcus aureus (MRSA): Antibiotic-resistance and the biofilm phenotype. Medchemcomm 2019, 10, 1231–1241. [Google Scholar] [CrossRef]
- Kortright, K.E.; Chan, B.K.; Koff, J.L.; Turner, P.E. Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria. Cell Host Microbe 2019, 25, 219–232. [Google Scholar] [CrossRef] [Green Version]
- Roy, B.; Ackermann, H.W.; Pandian, S.; Picard, G.; Goulet, J. Biological inactivation of adhering Listeria monocytogenes by listeriaphages and a quaternary ammonium compound. Appl. Environ. Microbiol. 1993, 59, 2914–2917. [Google Scholar] [CrossRef] [Green Version]
- Doolittle, M.M.; Cooney, J.J.; Caldwell, D.E. Tracing the interaction of bacteriophage with bacterial biofilms using fluorescent and chromogenic probes. J. Ind. Microbiol. 1996, 16, 331–341. [Google Scholar] [CrossRef]
- Hughes, K.A.; Sutherland, I.W.; Jones, M.V. Biofilm susceptibility to bacteriophage attack: The role of phage-borne polysaccharide depolymerase. Microbiology 1998, 144, 3039–3047. [Google Scholar] [CrossRef] [Green Version]
- Hanlon, G.W. Bacteriophages: An appraisal of their role in the treatment of bacterial infections. Int. J. Antimicrob. Agents 2007, 30, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Moye, Z.D.; Woolston, J.; Sulakvelidze, A. Bacteriophage applications for food production and processing. Viruses 2018, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; Shen, M.; Jiang, X.; Shen, W.; Zhong, Q.; Yang, Y.; Tan, Y.; Agnello, M.; He, X.; Hu, F.; et al. Transcriptomic and metabolomics profiling of phage-host interactions between phage PaP1 and Pseudomonas aeruginosa. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Smet, J.; Zimmermann, M.; Kogadeeva, M.; Ceyssens, P.J.; Vermaelen, W.; Blasdel, B.; Jang, H.B.; Sauer, U.; Lavigne, R. High coverage metabolomics analysis reveals phage-specific alterations to Pseudomonas aeruginosa physiology during infection. ISME J. 2016, 10, 1823–1835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández, L.; Gutiérrez, D.; Rodríguez, A.; García, P. Application of Bacteriophages in the Agro-Food Sector: A Long Way Toward Approval. Front. Cell. Infect. Microbiol. 2018, 8, 296. [Google Scholar] [CrossRef] [Green Version]
- Qian, W.; Jia, Y.; Ren, S.-X.; He, Y.-Q.; Feng, J.-X.; Lu, L.-F.; Sun, Q.; Ying, G.; Tang, D.-J.; Tang, H.; et al. Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv. campestris. Genome Res. 2005, 15, 757–767. [Google Scholar] [CrossRef] [Green Version]
- Dow, J.M.; Daniels, M.J. Pathogenicity determinants and global regulation of pathogenicity of Xanthomonas campestris pv. campestris. Curr. Top. Microbiol. Immunol. 1994, 192, 29–41. [Google Scholar]
- Liao, C.-T.; Chiang, Y.-C.; Hsiao, Y.-M. Functional characterization and proteomic analysis of lolA in Xanthomonas campestris pv. campestris. BMC Microbiol. 2019, 19, 20. [Google Scholar] [CrossRef] [Green Version]
- Fulgione, A.; Ianniello, F.; Papaianni, M.; Contaldi, F.; Sgamma, T.; Giannini, C.; Pastore, S.; Velotta, R.; Ventura, B.D.; Roveri, N.; et al. Biomimetic hydroxyapatite nanocrystals are an active carrier for Salmonella bacteriophages. Int. J. Nanomed. 2019, 14, 2219–2232. [Google Scholar] [CrossRef] [Green Version]
- Papa, R.; Selan, L.; Parrilli, E.; Tilotta, M.; Sannino, F.; Feller, G.; Tutino, M.L.; Artini, M. Anti-Biofilm Activities from Marine Cold Adapted Bacteria Against Staphylococci and Pseudomonas aeruginosa. Front. Microbiol. 2015, 6, 1333. [Google Scholar] [CrossRef] [Green Version]
- Papa, R.; Parrilli, E.; Sannino, F.; Barbato, G.; Tutino, M.L.; Artini, M.; Selan, L. Anti-biofilm activity of the Antarctic marine bacterium Pseudoalteromonas haloplanktis TAC125. Res. Microbiol. 2013, 164, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Casillo, A.; Casillo, A.; Parrilli, E.; Filomena, S.; Lindner, B.; Lanzetta, R.; Parrilli, M.; Tutino, M.L.; Corsaro, M.M. Structural Investigation of the Oligosaccharide Portion Isolated from the Lipooligosaccharide of the Permafrost Psychrophile Psychrobacter arcticus 273-4. Mar. Drugs 2015, 13, 4539–4555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papaianni, M.; Contaldi, F.; Fulgione, A.; Woo, S.L.; Casillo, A.; Corsaro, M.M.; Parrilli, E.; Marcolungo, L.; Rossato, M.; Delledonne, M.; et al. Role of phage ϕ1 in two strains of Salmonella Rissen, sensitive and resistant to phage ϕ1. BMC Microbiol. 2018, 18, 208. [Google Scholar] [CrossRef] [PubMed]
- Hwang, T.L.; Shaka, A.J. Water Suppression That Works. Excitation Sculpting Using Arbitrary Wave-Forms and Pulsed-Field Gradients. J. Magn. Reson. Ser. A 1995, 112, 275–279. [Google Scholar] [CrossRef]
- Bax, A.; Davis, D.G. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 1985, 65, 355–360. [Google Scholar] [CrossRef]
- Griesinger, C.; Otting, G.; Wuethrich, K.; Ernst, R.R. Clean TOCSY for proton spin system identification in macromolecules. J. Am. Chem. Soc. 1988, 110, 7870–7872. [Google Scholar] [CrossRef]
- Schleucher, J.; Schwendinger, M.; Sattler, M.; Schmidt, P.; Schedletzky, O.; Glaser, S.J.; Sørensen, O.W.; Griesinger, C. A general enhancement scheme in heteronuclear multidimensional NMR employing pulsed field gradients. J. Biomol. NMR 1994, 4, 301–306. [Google Scholar] [CrossRef]
- Trygg, J.; Wold, S. Orthogonal projections to latent structures (O-PLS). J. Chemom. 2002, 16, 119–128. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [Green Version]
- Walton, J.T.; Hill, D.J.; Protheroe, R.G.; Nevill, A.; Gibson, H. Investigation into the effect of detergents on disinfectant susceptibility of attached Escherichia coli and Listeria monocytogenes. J. Appl. Microbiol. 2008, 105, 309–315. [Google Scholar] [CrossRef]
- Casillo, A.; Papa, R.; Ricciardelli, A.; Sannino, F.; Ziaco, M.; Tilotta, M.; Selan, L.; Marino, G.; Corsaro, M.M.; Tutino, M.L.; et al. Anti-Biofilm Activity of a Long-Chain Fatty Aldehyde from Antarctic Pseudoalteromonas haloplanktis TAC125 against Staphylococcus epidermidis Biofilm. Front. Cell. Infect. Microbiol. 2017, 7, 46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.H.; Kim, Y.G.; Park, J.G.; Lee, J. Supercritical fluid extracts of Moringa oleifera and their unsaturated fatty acid components inhibit biofilm formation by Staphylococcus aureus. Food Control 2017, 80, 74–82. [Google Scholar] [CrossRef]
- Yoon, B.K.; Jackman, J.A.; Valle-González, E.R.; Cho, N.J. Antibacterial free fatty acids and monoglycerides: Biological activities, experimental testing, and therapeutic applications. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bravo-Santano, N.; Ellis, J.K.; Calle, Y.; Keun, H.C.; Behrends, V.; Letek, M. Intracellular staphylococcus aureus elicits the production of host very long-chain saturated fatty acids with antimicrobial activity. Metabolites 2019, 9. [Google Scholar] [CrossRef] [Green Version]
- Lade, H.; Park, J.H.; . Chung, S.H.; Kim, H.I.; Kim, J.-M.; Joo, H.-S.; Kim, J.-S. Biofilm Formation by Staphylococcus aureus Clinical Isolates is Differentially Affected by Glucose and Sodium Chloride Supplemented Culture Media. J. Clin. Med. 2019, 8, 1853. [Google Scholar] [CrossRef] [Green Version]
- Wong, H.S.; Maker, G.L.; Trengove, R.D.; O’Handley, R.M. Gas chromatography-mass spectrometry-based metabolite profiling of Salmonella enterica serovar typhimurium differentiates between biofilm and planktonic phenotypes. Appl. Environ. Microbiol. 2015, 81, 2660–2666. [Google Scholar] [CrossRef] [Green Version]
- Santi, L.; Beys-Da-Silva, W.O.; Berger, M.; Calzolari, D.; Guimarães, J.A.; Moresco, J.J.; Yates, J.R. Proteomic profile of Cryptococcus neoformans biofilm reveals changes in metabolic processes. J. Proteome Res. 2014, 13, 1545–1559. [Google Scholar] [CrossRef]
- Goodwine, J.; Gil, J.; Doiron, A.; Valdes, J.; Solis, M.; Higa, A.; Davis, S.; Sauer, K. Pyruvate-depleting conditions induce biofilm dispersion and enhance the efficacy of antibiotics in killing biofilms in vitro and in vivo. Sci. Rep. 2019, 9, 3763. [Google Scholar] [CrossRef] [Green Version]
- Ene, I.V.; Heilmann, C.J.; Sorgo, A.G.; Walker, L.A.; de Koster, C.G.; Munro, C.A.; Klis, F.M.; Brown, A.J.P. Carbon source-induced reprogramming of the cell wall proteome and secretome modulates the adherence and drug resistance of the fungal pathogen Candida albicans. Proteomics 2012, 12, 3164–3179. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Okuno, Y.; Cavagnero, S. Sensitivity enhancement in solution NMR: Emerging ideas and new frontiers. J. Magn. Reson. 2014, 241, 18–31. [Google Scholar] [CrossRef] [Green Version]
- Dubois-Brissonnet, F.; Trotier, E.; Briandet, R. The biofilm lifestyle involves an increase in bacterial membrane saturated fatty acids. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flemming, H.C.; Wingender, J. Relevance of microbial extracellular polymeric substances (EPSs) - Part I: Structural and ecological aspects. Water Sci. Technol. 2001, 43, 1–8. [Google Scholar] [CrossRef]
- Dea, I.C.M.; Morris, E.R.; Rees, D.A.; Welsh, E.J.; Barnes, H.A.; Price, J. Associations of like and unlike polysaccharides: Mechanism and specificity in galactomannans, interacting bacterial polysaccharides, and related systems. Carbohydr. Res. 1977, 57, 249–272. [Google Scholar] [CrossRef]
- Penner, J.C.; Ferreira, J.A.G.; Secor, P.R.; Sweere, J.M.; Birukova, M.K.; Joubert, L.-M.; Haagensen, J.A.J.; Garcia, O.; Malkovskiy, A.V.; Kaber, G.; et al. Pf4 bacteriophage produced by Pseudomonas aeruginosa inhibits Aspergillus fumigatus metabolism via iron sequestration. Microbiology 2016, 162, 1583–1594. [Google Scholar] [CrossRef] [PubMed]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papaianni, M.; Cuomo, P.; Fulgione, A.; Albanese, D.; Gallo, M.; Paris, D.; Motta, A.; Iannelli, D.; Capparelli, R. Bacteriophages Promote Metabolic Changes in Bacteria Biofilm. Microorganisms 2020, 8, 480. https://doi.org/10.3390/microorganisms8040480
Papaianni M, Cuomo P, Fulgione A, Albanese D, Gallo M, Paris D, Motta A, Iannelli D, Capparelli R. Bacteriophages Promote Metabolic Changes in Bacteria Biofilm. Microorganisms. 2020; 8(4):480. https://doi.org/10.3390/microorganisms8040480
Chicago/Turabian StylePapaianni, Marina, Paola Cuomo, Andrea Fulgione, Donatella Albanese, Monica Gallo, Debora Paris, Andrea Motta, Domenico Iannelli, and Rosanna Capparelli. 2020. "Bacteriophages Promote Metabolic Changes in Bacteria Biofilm" Microorganisms 8, no. 4: 480. https://doi.org/10.3390/microorganisms8040480
APA StylePapaianni, M., Cuomo, P., Fulgione, A., Albanese, D., Gallo, M., Paris, D., Motta, A., Iannelli, D., & Capparelli, R. (2020). Bacteriophages Promote Metabolic Changes in Bacteria Biofilm. Microorganisms, 8(4), 480. https://doi.org/10.3390/microorganisms8040480