Antimicrobial Peptides and Physical Activity: A Great Hope against COVID 19
Abstract
:1. Introduction
2. Characteristics of Coronavirus
SARS-Cov-2 Infection
3. Defensins: Alpha and Beta-Defensins
4. LL37: Human Cathelicidin
5. Defensins against Coronaviruses
6. Mechanisms of Action of Defensins
6.1. Mouse β-Defensins-4 Derived P9
6.2. HD5: Human Intestinal α-Defensin-5
6.3. HBD2: Human Beta Defensin 2
6.4. RTD-1: Rhesus θ-Defensin 1
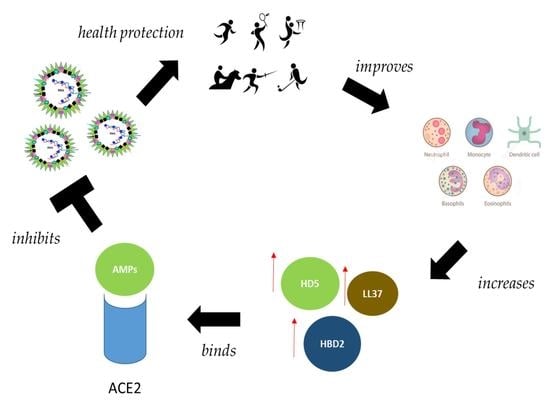
7. Interplay between Physical Activity, Human Defensins and Immune System
8. Vitamin D Generates Defensins Active against COVID-19
9. Other Therapeutic Defensins
10. Defensins as Vaccine Adjuvants
11. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Brice, D.C.; Diamond, G. Antiviral activities of human host defense peptides. Curr. Med. Chem. 2020, 27, 1420–1443. [Google Scholar] [CrossRef]
- Hancock, R.E.; Haney, E.F.; Gill, E.E. The immunology of host defence peptides: Beyond antimicrobial activity. Nat. Rev. Immunol. 2016, 16, 321–334. [Google Scholar] [CrossRef]
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug Discov. 2020, 19, 311–332. [Google Scholar] [CrossRef] [PubMed]
- Angrisano, T.; Pero, R.; Paoletti, I.; Keller, S.; Lembo, L.; Baroni, A.; Chiariotti, L.; Lembo, F.; Donnarumma, G. Epigenetic regulation of IL-8 and β-defensin genes in human keratinocytes in response to Malassezia furfur. J. Investig. Dermatol. 2013, 133, 2101–2104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coretti, L.; Natale, A.; Cuomo, M.; Florio, E.; Keller, S.; Lembo, F.; Chiariotti, L.; Pero, R. The Interplay between Defensins and Microbiota in Crohn’s Disease. Mediat. Inflamm. 2017, 2017, 8392523. [Google Scholar] [CrossRef] [PubMed]
- Pero, R.; Coretti, L.; Nigro, E.; Lembo, F.; Laneri, S.; Lombardo, B.; Daniele, A.; Scudiero, O. Defensins in the Fight against Helicobacter pylori. Molecules 2017, 22, 424. [Google Scholar] [CrossRef] [PubMed]
- Pero, R.; Brancaccio, M.; Laneri, S.; Biasi, M.G.; Lombardo, B.; Scudiero, O. A Novel View of Human Helicobacter pylori Infections: Interplay between Microbiota and Beta-Defensins. Biomolecules 2019, 9, 237. [Google Scholar] [CrossRef] [Green Version]
- Pero, R.; Angrisano, T.; Brancaccio, M.; Falanga, A.; Lombardi, L.; Natale, F.; Laneri, S.; Lombardo, B.; Galdiero, S.; Scudiero, O. Beta-defensins and analogs in Helicobacter pylori infections: mRNA expression levels, DNA methylation, and antibacterial activity. PLoS ONE 2019, 14, e0222295. [Google Scholar] [CrossRef] [Green Version]
- Pero, R.; Brancaccio, M.; Mennitti, C.; Gentile, L.; Franco, A.; Laneri, S.; De Biasi, M.G.; Pagliuca, C.; Colicchio, R.; Salvatore, P.; et al. HNP-1 and HBD-1 as Biomarkers for the Immune Systems of Elite Basketball Athletes. Antibiotics 2020, 9, 306. [Google Scholar] [CrossRef]
- Scudiero, O.; Brancaccio, M.; Mennitti, C.; Laneri, S.; Lombardo, B.; De Biasi, M.G.; De Gregorio, E.; Pagliuca, C.; Colicchio, R.; Salvatore, P.; et al. Human Defensins: A Novel Approach in the Fight against Skin Colonizing Staphylococcus aureus. Antibiotics 2020, 9, 198. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, A.; Siman-Tov, G.; Hall, G.; Bhalla, N.; Narayanan, A. Human antimicrobial peptides as therapeutics for viral infections. Viruses 2019, 11, 704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doss, M.; White, M.R.; Tecle, T.; Hartshorn, K.L. Human defensins and LL-37 in mucosalimmunity. J. Leukoc. Biol. 2010, 87, 79–92. [Google Scholar] [CrossRef]
- Prasad, S.V.; Fiedoruk, K.; Daniluk, T.; Piktel, E.; Bucki, R. Expression and function of host defense peptides at inflammation sites. Int. J. Mol. Sci. 2019, 21, 104. [Google Scholar] [CrossRef] [Green Version]
- Kudryashova, E.; Zani, A.; Vilmen, G.; Sharma, A.; Lu, W.; Yount, J.S.; Kudryashov, D.S. SARS-CoV-2 incativation by human defensin HNP1 and retrocyclin RC-101. bioRxiv 2021. [Google Scholar] [CrossRef]
- Idris, M.M.; Banu, S.; Siva, A.B.; Nagaraj, R. Downregulation of Defensin genes in SARS-CoV-2 infection. medRxiv 2021. [Google Scholar] [CrossRef]
- Solanki, S.S.; Singh, P.; Kashyap, P.; Sansi, M.S.; Ali, S.A. Promising role of defensins peptides as therapeutics to combat against viral infection. Microb. Pathog. 2021, 155, 104930. [Google Scholar] [CrossRef] [PubMed]
- Davison, G.; Kehaya, C.; Wyn Jones, A. Nutritional and Physical Activity Interventions to Improve Immunity. Am. J. Lifestyle Med. 2014, 10, 152–169. [Google Scholar] [CrossRef] [Green Version]
- Brancaccio, M.; Mennitti, C.; Laneri, S.; Franco, A.; De Biasi, M.G.; Cesaro, A.; Fimiani, F.; Moscarella, E.; Gragnano, F.; Mazzaccara, C.; et al. Methicillin-Resistant Staphylococcus aureus: Risk for General Infection and Endocarditis Among Athletes. Antibiotics 2020, 9, 332. [Google Scholar] [CrossRef]
- Brancaccio, M.; Mennitti, C.; Cesaro, A.; Fimiani, F.; Moscarella, E.; Caiazza, M.; Gragnano, F.; Ranieri, A.; D’Alicandro, G.; Tinto, N.; et al. Dietary Thiols: A Potential Supporting Strategy against Oxidative Stress in Heart Failure and Muscular Damage during Sports Activity. Int. J. Environ. Res. Public Health 2020, 17, 9424. [Google Scholar] [CrossRef] [PubMed]
- Brancaccio, M.; Mennitti, C.; Gentile, A.; Correale, L.; Buzzachera, C.F.; Ferraris, C.; Montomoli, C.; Frisso, G.; Borrelli, P.; Scudiero, O. Effects of the COVID-19 Pandemic on Job Activity, Dietary Behaviours and Physical Activity Habits of University Population of Naples, Federico II-Italy. Int. J. Environ. Res. Public Health 2021, 18, 1502. [Google Scholar] [CrossRef]
- Romeo, J.; Wärnberg, J.; Pozo, T.; Marcos, A. Physical activity, immunity and infection. Proc. Nutr. Soc. 2010, 69, 390–399. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Morales, A.J.; Bonilla-Aldana, D.K.; Balbin-Ramon, G.J.; Rabaan, A.A.; Sah, R.; Paniz-Mondolfi, A.; Pagliano, P.; Esposito, S. History is repeating itself: Probable zoonotic spillover as the cause of the 2019 novel Coronavirus Epidemic. Infez. Med. 2020, 28, 3–5. [Google Scholar] [PubMed]
- Rothan, H.A.; Byrareddy, S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020, 109, 102433. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Laboratory Testing of Human Suspected Cases of Novel Coronavirus (nCoV) Infection. Available online: https://www.who.int/publications/i/item/10665-330374 (accessed on 10 January 2020).
- World Health Organization. Novel Coronavirus (2019-nCoV) Situation Report-2. Available online: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200122-sitrep-2-2019-ncov.pdf?sfvrsn=4d5bcbca_2 (accessed on 22 January 2020).
- World Health Organization. Middle East Respiratory Syndrome Coronavirus (MERS-CoV). Available online: https://www.who.int/health-topics/middle-east-respiratory-syndrome-coronavirus-mers#tab=tab_1 (accessed on 19 January 2020).
- World Health Organization. WHO MERS Global Summary and Assessment of Risk. Available online: https://apps.who.int/iris/bitstream/handle/10665/326126/WHO-MERS-RA-19.1-eng.pdf?ua=1 (accessed on 19 July 2019).
- Malik, Y.S.; Kumar, N.; Sircar, S.; Kaushik, R.; Bhat, S.; Dhama, K.; Gupta, P.; Goyal, K.; Singh, M.P.; Ghoshal, U.; et al. Coronavirus Disease Pandemic (COVID-19): Challenges and a Global Perspective. Pathogens 2020, 9, 519. [Google Scholar] [CrossRef]
- Li, G.; Fan, Y.; Lai, Y.; Han, T.; Li, Z.; Zhou, P.; Pan, P.; Wang, W.; Hu, D.; Liu, X.; et al. Coronavirus infections and immune responses. J. Med. Virol. 2020, 92, 424–432. [Google Scholar] [CrossRef]
- Scudiero, O.; Lombardo, B.; Brancaccio, M.; Mennitti, C.; Cesaro, A.; Fimiani, F.; Gentile, L.; Moscarella, E.; Amodio, F.; Ranieri, A.; et al. Exercise, Immune System, Nutrition, Respiratory and Cardiovascular Diseases during COVID-19: A Complex Combination. Int. J. Environ. Res. Public Health 2021, 18, 904. [Google Scholar] [CrossRef]
- Słomka, A.; Kowalewski, M.; Żekanowska, E. Coronavirus Disease 2019 (COVID-19): A Short Review on Hematological Manifestations. Pathogens 2020, 9, 493. [Google Scholar] [CrossRef]
- Gencer, S.; Lacy, M.; Atzler, D.; van der Vorst, E.P.C.; Döring, Y.; Weber, C. Immunoinflammatory, Thrombohaemostatic, and Cardiovascular Mechanisms in COVID-19. Thromb. Haemost. 2020, 120, 1629–1641. [Google Scholar] [CrossRef]
- Rabi, F.A.; Al Zoubi, M.S.; Kasasbeh, G.A.; Salameh, D.M.; Al-Nasser, A.D. SARS-CoV-2 and Coronavirus Disease 2019: What We Know So Far. Pathogens 2020, 9, 231. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.-L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, S.; Zhu, Y.; Liu, M.; Lan, Q.; Xu, W.; Wu, Y. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell. Mol. Immunol. 2020, 17, 765–767. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef]
- Wölfel, R.; Victor, M.C.; Wolfgang, G.; Michael, S.; Sabine, Z.; Marcel, A.M.; Daniela, N.; Terry, C.J.; Patrick, V.; Camilla, R.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziegler, C.G.K.; Allons, S.J.; Nyquist, S.K.; Mbano, I.M.; Miao, V.N.; Tzounanas, C.N.; Cao, Y.; Yousif, A.S.; Bals, J.; Hauser, B.M.; et al. SARS- CoV-2 receptor ACE2 is an interferon- stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 2020, 181, 1016–1035. [Google Scholar] [CrossRef]
- Blanco-Melo, D.; Nillson-Payant, B.E.; Liu, W.-C.; Uhl, S.; Hoagland, D.; Moller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced host response to SARS- CoV-2 drives development of COVID-19. Cell 2020. [Google Scholar] [CrossRef]
- Tortorici, M.A.; Veesler, D. Structural insights into coronavirus entry. Adv. Virus Res. 2019, 105, 93–116. [Google Scholar]
- Li, F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef] [Green Version]
- Letko, M.; Marzi, A.; Munster, V. Functional assessment of cell entry and receptor usage for SARS- CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. [Google Scholar] [CrossRef] [Green Version]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterization of spike glycoprotein of SARS- CoV-2 on virus entry and its immunecross- reactivity with SARS- CoV. Nat. Commun. 2020, 11, 1620. [Google Scholar] [CrossRef] [Green Version]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS- CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef] [PubMed]
- Menachery, V.D.; Dinnon, K.H., 3rd; Yount, B.L., Jr.; McAnarney, E.T.; Gralinski, L.E.; Hale, A.; Graham, R.L.; Scobey, T.; Anthony., S.-J.; Wang, L.; et al. Trypsin treatment unlocks barrier for zoonotic bat coronavirus infection. J. Virol. 2020, 94, e01774-19. [Google Scholar] [CrossRef] [Green Version]
- Coutard, B.; Valle, C.; de Lamballerie, X.; Canard, B.; Seidah, N.G.; Decroly, E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin- like cleavage site absent in CoV of the same clade. Antivir. Res. 2020, 176, 104742. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, K.-W.; Pöhlmann, S. A multibasic cleavage site in the spike protein of SARS- CoV-2 is essential for infection of human lung cells. Mol. Cell 2020, 78, 779–784.e5. [Google Scholar] [CrossRef]
- Buchholz, U.J.; Bukreyev, A.; Yang, L.; Lamirande, E.W.; Murphy, B.R.; Subbarao, K.; Collins, P.L. Contributions of the structural proteins of severe respiratory syndrome coronavirus to protective immunity. Proc. Natl. Acad. Sci. USA 2004, 101, 9804–9809. [Google Scholar] [CrossRef] [Green Version]
- Walls, A.C.; Tortorici, M.A.; Frenz, B.; Snijder, J.; Li, W.; Rey, F.A.; DIMaio, F.; Bosch, B.-J.; Veesler, D. Glycan shield and epitope masking of a coronavirus spike protein observed by cryoelectron microscopy. Nat. Struct. Mol. Biol. 2016, 23, 899–905. [Google Scholar] [CrossRef]
- Watanabe, Y.; Allen, J.D.; Wrapp, D.; McLellan, J.S.; Crispin, M. Site- specific glycan analysis of the SARS- CoV-2 spike. Science 2020. [Google Scholar] [CrossRef] [PubMed]
- Pinto, D.; Park, Y.-J.; Beltramello, M.; Walls, A.C.; Tortorici, M.A.; Bianchi, S.; Jaconi, S.; Culap, K.; Zatta, F.; De Marco, A.; et al. Cross- neutralization of SARS- CoV-2 by a human monoclonal SARS- CoV antibody. Nature 2020, 583, 290–295. [Google Scholar] [CrossRef]
- Wang, C.; Li, W.; Drabek, D.; Okba, N.M.A.; Haperen, R.; Oster, A.D.M.E.; van Kupperveld, F.J.M.; Haagmans, B.L.; Grosveld, F.; Bosh, B.-J. A human monoclonal antibody blocking SARS- CoV-2 infection. Nat. Commun. 2020, 11, 2251. [Google Scholar] [CrossRef]
- Cao, Y.; Su, B.; Guo, X.; Sun, W.; Deng, Y.; Bao, L.; Zhu, Q.; Zhang, X.; Zheng, Y.; Geng, C.; et al. Potent neutralizing antibodies against SARS- CoV-2 identified by high- throughput single- cell sequencing of convalescent patients’ B cells. Cell 2020, 182, 73–84. [Google Scholar] [CrossRef]
- Wrapp, D.; De Vlieger, D.; Corbett, K.S.; Torres, G.M.; Wang, N.; Van Breedam, W.; Roose, K.; van Schie, L.; VIB-CMB COVID-19 Response Team; Hoffmann, M.; et al. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell 2020, 181, 1436–1441. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, F.; Shen, C.; Peng, W.; Li, D.; Zhao, C.; Li, Z.; Li, S.; Bi, Y.; Yang, Y.; et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science 2020, 368, 1274–1278. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Xiao, G.; Chen, Y.; He, Y.; Niu, J.; Escalante, C.R.; Xiong, H.; Farmar, J.; Debnath, A.K.; Tien, P.; et al. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: Implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet 2004, 363, 938–947. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.; Peng, W.; Zhu, Y.; Lu, S.; Zhou, M.; Lin, W.; Wu, W.; Huang, S.; Jiang, L.; Luo, X.; et al. Recent progress in understanding 2019 novel coronavirus (SARS-CoV-2) associated with human respiratory disease: Detection, mechanisms and treatment. Int. J. Antimicrob. Agents 2020, 55, 1059. [Google Scholar] [CrossRef]
- Du, L.; He, Y.; Zhou, Y.; Liu, S.; Zheng, B.J.; Jiang, S. The spike protein of SARS-CoV—A target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2020, 7, 226–236. [Google Scholar] [CrossRef]
- Das, K.; Aramini, J.M.; Ma, L.C.; Krug, R.M.; Arnold, E. Structures of influenza A proteins and insights into antiviral drug targets. Nat. Struct. Mol. Biol. 2010, 17, 530–538. [Google Scholar] [CrossRef] [Green Version]
- Gorbalenya, A.E.; Enjuanes, L.; Ziebuhr, J.; Snijder, E.J. Nidovirales: Evolving the largest RNA virus genome. Virus Res. 2006, 117, 17–37. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Dellibovi-Ragheb, T.A.; Pak, E.; Qiu, Q.; Fisher, M.; Takvorian, P.M.; Bleck, C.; Hsu, V.; Fehr, A.R.; Perlman, S.; et al. β-Coronaviruses use lysosomal organelles for cellular egress. bioRxiv 2020. [Google Scholar] [CrossRef]
- Kim, D.; Lee, J.-Y.; Yang, J.-S.; Kim, J.W.; Kim, V.N.; Chang, H. The architecture of SARS- CoV-2 transcriptome. Cell 2020, 181, 914–921. [Google Scholar] [CrossRef]
- McIntosh, K.; Dees, J.H.; Becker, W.B.; Kapikian, A.Z.; Chanock, R.M. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc. Natl. Acad. Sci. USA 1967, 57, 933–940. [Google Scholar] [CrossRef] [Green Version]
- Xu, K.; Zheng, B.J.; Zeng, R.; Lu, W.; Lin, Y.P.; Xue, L.; Li, L.; Yang, L.L.; Xu, C.; Dai, J.; et al. Severe acute respiratory syndrome coronavirus accessory protein 9b is a virion- associated protein. Virology 2009, 388, 279–285. [Google Scholar] [CrossRef]
- Davidson, A.D.; Williamson, M.K.; Lewis, S.; Shoemark, D.; Carrol, M.W.; Heesom, K.; Zambon, M.; Ellis, J.; Lewis, P.A.; Hiscox, J.A.; et al. Characterisation of the transcriptome and proteome of SARS- CoV-2 reveals a cell passage induced in- frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 2020, 12, 68. [Google Scholar] [CrossRef] [PubMed]
- Perlman, S.; Netland, J. Coronaviruses post- SARS: Update on replication and pathogenesis. Nat. Rev. Microbiol. 2009, 7, 439–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masters, P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006, 65, 193–292. [Google Scholar]
- Hosoki, K.; Chakraborty, A.; Sur, S. Molecular mechanisms and epidemiology of COVID-19 from an allergist’s perspective. J. Allergy Clin. Immunol. 2020, 146, 285–299. [Google Scholar] [CrossRef]
- Liu, D.X.; Fung, T.S.; Chong, K.K.L.; Shukla, A.; Hilgenfeld, R. Accessory proteins of SARS- CoV and other coronaviruses. Antivir. Res. 2014, 109, 97–109. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J.; Chen, J.; Luo, B.; Yuan, Y.; Huang, F.; Yang, T.; Yu, F.; Liu, J.; Liu, B.; et al. The ORF8 protein of SARS- CoV-2 mediates immune evasion through potently downregulating MHC- I. bioRxiv 2020. [Google Scholar] [CrossRef]
- Patil, A.; Hughes, A.L.; Zhang, G. Rapid evolution and diversification of mammalian α-defensins as revealed by comparative analysis of rodent and primate genes. Physiol. Genom. 2004, 20, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Cunliffe, R.N. α-Defensins in the gastrointestinal tract. Mol. Immunol. 2003, 7, 463–467. [Google Scholar] [CrossRef]
- Lehrer, R.I. Primate defensins. Nat. Rev. Microbiol. 2004, 9, 727–738. [Google Scholar] [CrossRef]
- Kawsar, H.I.; Ghosh, S.K.; Hirsch, S.A.; Koon, H.B.; Weinberg, A.; Jin, G. Expression of human beta-defensin-2 in intratumoral vascular endothelium and in endothelial cells induced by transforming growth factor beta. Peptides 2010, 31, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Ryan, L.K.; Dai, J.; Yin, Z.; Megjugorac, N.; Uhlhorn, V.; Yim, S.; Schwartz, K.D.; Abrahams, J.A.; Diamond, G.; Fitzgerald-Bocarsly, P. Modulation of human beta-defensin-1 (hBD-1) inplasmacytoid dendritic cells (PDC), monocytes, and epithelial cells by influenza virus, Herpes simplex virus, and Sendai virus and its possible role in innate immunity. J. Leukoc. Biol. 2011, 90, 343–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehlotra, R.K.; Zimmerman, P.A.; Weinberg, A. Defensin gene variation and HIV/AIDS: A comprehensive perspective needed. J. Leukoc. Biol. 2016, 99, 687–692. [Google Scholar] [CrossRef] [Green Version]
- Chessa, C.; Bodet, C.; Jousselin, C.; Wehbe, M.; Leveque, N.; Garcia, M. Antiviral and immunomodulatory properties of antimicrobial peptides produced by human keratinocytes. Front. Microbiol. 2020, 11, 1155. [Google Scholar] [CrossRef]
- Park, M.S.; Kim, J.I.; Lee, I.; Park, S.; Bae, J.Y.; Park, M.S. Towards the application of human defensins as antivirals. Biomol. Ther. 2018, 26, 242–254. [Google Scholar] [CrossRef]
- Bjorstad, A.; Askarieh, G.; Brown, K.L.; Chrisetnson, K.; Forsman, H.; Onnheim, K.; Li, H.-M.; Teneberg, S.; Maier, O.; Hoekstra, D.; et al. The host defense peptide LL-37 selectively permeabilizes apoptotic leukocytes. Antimicrob. Agents Chemother. 2009, 53, 1027–1038. [Google Scholar] [CrossRef] [Green Version]
- Sorensen, O.E.; Follin, P.; Johnsen, A.H.; Follin, P.; Johnsen, A.H.; Tjabringa, G.S.; Hiemstra, P.S.; Boregaard, N. Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3. Blood 2001, 97, 3951–3959. [Google Scholar] [CrossRef] [Green Version]
- Barlow, P.G.; Svoboda, P.; Mackellar, A.; Nash, A.A.; York, I.A.; Pohl, J.; Davidson, D.J. Antiviral activity and increased host defense against influenza infection elicited by the human cathelicidin LL-37. PLoS ONE 2011, 6, e25333. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhao, Y.; Jiang, X.; Zhao, Y.; Yang, L.; Chen, L.; Dong, M.; Luan, Z.; Chunlong, Y.; Jiao, J.; et al. Preliminary evaluation of the safety and efficacy of oral human antimicrobial peptide LL-37 in the treatment of patients of COVID-19, a small-scale, single-arm, exploratory safety. medRxiv 2020. [Google Scholar] [CrossRef]
- Lokhande, K.B.; Banerjee, T.; Swamy, K.V.; Deshpande, M. An in silico scientific basis for LL-37 as a therapeutic and vitamin D as preventive for Covid-19. ChemRxiv 2020. [Google Scholar] [CrossRef]
- Wang, C.; Wang, S.; Li, D.; Chen, P.; Han, S.; Zhao, G.; Chen, Y.; Zhao, J.; Xiong, J.; Qiu, J.; et al. Human Cathelicidin Inhibits SARS-CoV-2 Infection: Killing Two Birds with One Stone. ACS Infect Dis. 2021. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, C.; Xu, X.F.; Xu, W.; Liu, S.W. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef] [PubMed]
- Whisenant, J.; Burgess, K. Blocking coronavirus 19 infection via the SARS-CoV-2 spike protein: Initial steps. ACS Med. Chem. Lett. 2020, 11, 1076–1078. [Google Scholar] [CrossRef]
- Wang, C.; Wang, S.; Li, D.; Wei, D.Q.; Zhao, J.; Wang, J. Human intestinal defensin 5 inhibits SARS-CoV-2 invasion by cloaking ACE2. Gastroenterology 2020, 159, 1145–1147.e4. [Google Scholar] [CrossRef]
- Zhang, L.; Ghosh, S.K.; Basavarajappa, S.C.; Muller-Greven, J.; Penfield, J.; Brewer, A.; Ramakrishnan, P.; Buck, M.; Weinberg, A. Molecular dynamics simulations and functional studies reveal that hBD-2 binds SARS-CoV-2 spike RBD and blocks viral entry into ACE2 expressing cells. bioRxiv 2021. [Google Scholar] [CrossRef]
- Venkataraman, N.; Cole, A.L.; Ruchala, P.; Waring, A.J.; Stuchlik, O.; Cole, A.M. Reawakening retrocyclins: Ancestral human defensins active against HIV-1. PLoS Biol. 2009, 7, e95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehrer, R.I.; Cole, A.M.; Selsted, M.E. θ-Defensins: Cyclic peptides with endless potential. J. Biol. Chem. 2012, 287, 27014–27019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chalichem, N.S.S.; Bethapudi, B.; Mundkinajeddu, D. Aminoglycosides can be a better choice over macrolides in COVID-19 regimen: Plausible mechanism for repurposing strategy. Med. Hypotheses 2020, 144, 109984. [Google Scholar] [CrossRef]
- Wang, T.T.; Nestel, F.P.; Bourdeau, V.; Nagai, Y.; Wang, Q.; Liao, J.; Tavera-Mendoza, L.; Lin, R.; Hanrahan, J.H.; Mader, S.; et al. Cutting Edge: 1,25-Dihydroxyvitamin D3 Is a Direct Inducer of Antimicrobial Peptide Gene Expression. J. Immunol. 2004, 173, 2909–2912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dixon, B.M.; Barker, T.; McKinnon, T.; Cuomo, J.; Frei, B.; Borregaard, N.; Gombart, A.F. Positive correlation between circulating cathelicidin antimicrobial peptide (hCAP18/LL-37) and 25-hydroxyvitamin D levels in healthy adults. BMC Res. Notes 2012, 5, 575. [Google Scholar] [CrossRef] [Green Version]
- Tecle, T.; Tripathi, S.; Hartshorn, K.L. Review: Defensins and cathelicidins in lung immunity. Innate Immun. 2010, 16, 151–159. [Google Scholar] [CrossRef]
- Kaufman, H.W.; Niles, J.K.; Kroll, M.H.; Bi, C.; Holick, M.F. SARS-CoV-2 positivity rates associated with circulating 25-hydroxyvitamin D levels. PLoS ONE 2020, 15, e0239252. [Google Scholar]
- Arvinte, C.; Singh, M.; Marik, P.E. Serum levels of vitamin C and vitamin D in a cohort of critically ill COVID-19 patients of a north American community hospital intensive care unit in May 2020: A pilot study. Med. Drug Discov. 2020, 8, 100064. [Google Scholar] [CrossRef]
- Maghbooli, Z.; Sahraian, M.A.; Ebrahimi, M.; Pazoki, M.; Kafan, S.; Tabriz, H.M.; Hadadi, A.; Montazeri, M.; Nasiri, M.; Shirvani, A.; et al. Vitamin D sufficiency, a serum 25- hydroxyvitamin D at least 30 ng/mL reduced risk for adverse clinical outcomes in patients with COVID-19 infection. PLoS ONE 2020, 15, e0239799. [Google Scholar] [CrossRef] [PubMed]
- Radujkovic, A.; Hippchen, T.; Tiwari-Heckler, S.; Dreher, S.; Boxberger, M.; Merle, U. Vitamin D deficiency and outcome of COVID-19 patients. Nutrients 2020, 12, 2757. [Google Scholar] [CrossRef] [PubMed]
- Crane-Godreau, M.A.; Clem, K.J.; Payne, P.; Fiering, S. Vitamin D deficiency and air pollution exacerbate COVID-19 through suppression of antiviral peptide LL37. Front. Public Health 2020, 8, 232. [Google Scholar] [CrossRef]
- Hernández, J.L.; Nan, D.; Fernandez-Ayala, M.; García-Unzueta, M.; Hernández-Hernández, M.A.; López-Hoyos, M.; Muñoz-Cacho, P.; Olmos, J.M.; Gutiérrez-Cuadra, M.; Ruiz-Cubillán, J.J.; et al. Vitamin D Status in Hospitalized Patients with SARS-CoV-2 Infection. J. Clin. Endocrinol. Metab. 2021, 106, e1343–e1353. [Google Scholar] [CrossRef]
- Memariani, H.; Memariani, M. Therapeutic and prophylactic potential of anti-microbial peptides against coronaviruses. Ir. J. Med. Sci. 2020, 189, 1153–1154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, H.P.; Wowk, S.A.; Schutte, B.C.; Lee, S.K.; Vivado, A.; Tack, B.F.; Bevins, C.L.; McCray, P.B., Jr. A novel murine beta-defensin expressed in tongue, esophagus, and trachea. J. Biol. Chem. 2000, 275, 33314–33320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mustafa, S.; Balkhy, H.; Gabere, M. Peptide-protein interaction studies of antimicrobial peptides targeting Middle East Respiratory Syndrome coronavirus spike protein: An in silico approach. Adv. Bioinform. 2019, 2019, 6815105. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhou, J.; Zhang, K.; Chu, H.; Liu, D.; Poon, V.K.; Chan, C.C.; Leung, H.C.; Fai, N.; Lin, Y.P.; et al. A novel peptide with potent and broad-spectrum antiviral activities against multiple respiratory viruses. Sci. Rep. 2016, 6, 22008. [Google Scholar] [CrossRef]
- Zhao, H.; To, K.K.W.; Sze, K.H.; Yung, T.T.; Bian, M.; Lam, H.; Yeung, M.L.; Li, C.; Chu, H.; Yuen, K.Y. A broad-spectrum virus- and host-targeting peptide against respiratory viruses including influenza virus and SARS-CoV-2. Nat. Commun. 2020, 11, 4252. [Google Scholar] [CrossRef]
- Wohlford-Lenane, C.L.; Meyerholz, D.K.; Perlman, S.; Zhou, H.; Tran, D.; Selsted, M.E.; McCray, P.B., Jr. Rhesus theta-defensin prevents death in a mouse model of severe acute respiratory syndrome coronavirus pulmonary disease. J. Virol. 2009, 83, 11385–11390. [Google Scholar] [CrossRef] [Green Version]
- Luan, J.; Ren, Y.; Gao, S.; Zhang, L. High level of defensin alpha 5 in intestine may explain the low incidence of diarrhoea in COVID-19 patients. Eur. J. Gastroenterol. Hepatol. 2020. [Google Scholar] [CrossRef]
- Diamond, G.; Beckloff, N.; Ryan, L.K. Host defense peptides in the oral cavity and the lung: Similarities and differences. J. Dent. Res. 2008, 87, 915–927. [Google Scholar] [CrossRef] [PubMed]
- Ooi, C.Y.; Pang, T.; Leach, S.T.; Katz, T.; Day, A.S.; Jaffe, A. Fecal Human β-Defensin 2 in Children with Cystic Fibrosis: Is There a Diminished Intestinal Innate Immune Response? Dig. Dis. Sci. 2015, 60, 2946–2952. [Google Scholar] [CrossRef] [PubMed]
- Rivas-Santiago, B.; Schwander, S.K.; Sarabia, C.; Diamond, G.; Klein-Patel, M.E.; Hernandez-Pando, R.; Ellner, J.J.; Sada, E. Human {beta}-defensin 2 is expressed and associated with Mycobacterium tuberculosis during infection of human alveolar epithelial cells. Infect. Immun. 2005, 73, 4505–4511. [Google Scholar] [CrossRef] [Green Version]
- Semple, F.; Dorin, J.R. β-Defensins: Multifunctional modulators of infection, inflammation and more? J. Innate Immun. 2012, 4, 337–348. [Google Scholar] [CrossRef]
- Kota, S.; Sabbah, A.; Chang, T.H.; Harnack, R.; Xiang, Y.; Meng, X.; Bose, S. Role of human beta-defensin-2 during tumor necrosis factor-alpha/NF-kappaB-mediated innate antiviral response against human respiratory syncytial virus. J. Biol. Chem. 2008, 283, 22417–22429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawai, M.V.; Jia, H.P.; Liu, L.; Aseyev, V.; Wiencek, J.M.; McCray, P.B., Jr.; Ganz, T.; Kearney, W.R.; Tack, B.F. The NMR structure of human beta-defensin-2 reveals a novel alpha-helical segment. Biochemistry 2001, 40, 3810–3816. [Google Scholar] [CrossRef]
- Yeasmin, R.; Buck, M.; Weinberg, A.; Zhang, L. Translocation of Human β Defensin Type 3 through a Neutrally Charged Lipid Membrane: A Free Energy Study. J. Phys. Chem. B 2018, 122, 11883–11894. [Google Scholar] [CrossRef]
- Barros, E.P.; Casalino, L.; Gaieb, Z.; Dommer, A.C.; Wang, Y.; Fallon, L.; Raguette, L.; Belfon, K.; Simmerling, C.; Amaro, R.E. The Flexibility of ACE2 in the Context of SARS-CoV-2 Infection. Biophys. J. 2021, 6, 1072–1084. [Google Scholar] [CrossRef] [PubMed]
- Ghorbani, M.; Brooks, B.R.; Klauda, J.B. Critical Sequence Hotspots for Binding of Novel Coronavirus to Angiotensin Converter Enzyme as Evaluated by Molecular Simulations. J. Phys. Chem. B 2020, 124, 10034–10047. [Google Scholar] [CrossRef]
- Spinello, A.; Saltalamacchia, A.; Magistrato, A. Is the Rigidity of SARS-CoV-2 Spike Receptor-Binding Motif the Hallmark for Its Enhanced Infectivity? Insights from All-Atom Simulations. J. Phys. Chem. Lett. 2020, 11, 4785–4790. [Google Scholar] [CrossRef]
- Gombart, A.F.; Borregaard, N.; Koeffler, H.P. Human cathelicidin antimicrobial peptide (CAMP) gene is a direct target of the vitamin D receptor and is strongly up-regulated in myeloid cells by 1,25-dihydroxyvitamin D3. FASEB J. 2005, 19, 1067–1077. [Google Scholar] [CrossRef] [Green Version]
- Weber, G.; Heilborn, J.D.; Chamorro Jimenez, C.I.; Hammarsjo, A.; Torma, H.; Stahle, M. Vitamin D induces the antimicrobial protein hCAP18 in human skin. J. Investig. Dermatol. 2005, 124, 1080–1082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, P.T.; Stenger, S.; Li, H.; Wenzel, L.; Tan, B.H.; Krutzik, S.R.; Ochoa, M.T.; Schauber, J.; Wu, K.; Meinken, C.; et al. Toll-like receptor triggering of a vitamin D Mediated human antimicrobial response. Science 2006, 311, 1770–1773. [Google Scholar] [CrossRef] [PubMed]
- Ginde, A.A.; Liu, M.C.; Camargo, C.A. Demographic Differences and Trends of Vitamin D Insufficiency in the US Population. 1988–2004. Arch. Intern. Med. 2009, 169, 626–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Baylin, A.; Levy, P.D. Vitamin D deficiency and insufficiency among US adults: Prevalence, predictors and clinical implications. Br. J. Nutr. 2018, 119, 928–936. [Google Scholar] [CrossRef] [Green Version]
- The Coronavirus Is Infecting and Killing Black Americans at an Alarmingly High Rate. Available online: https://www.washingtonpost.com/nation/2020/04/07/coronavirus-is-infectingkillingblackamericans-an-alarmingly-high-rate-post-analysis-shows/?arc404=true (accessed on 7 April 2020).
- Virus Is Twice as Deadly for Black and Latino People Than Whites in N.Y.C. Available online: https://www.nytimes.com/2020/04/08/nyregion/coronavirus-race-deaths.html (accessed on 8 April 2020).
- Khare, D.; Godbole, N.M.; Pawar, S.D.; Mohan, V.; Pandey, G.; Gupta, S.; Kumar, D.; Dhole, T.P.; Godbole, M.M. Calcitriol [1, 25[OH]2 D3] pre- and post-treatment suppresses inflammatory response to influenza A (H1N1) infection in human lung A549 epithelial cells. Eur. J. Nutr. 2013, 52, 1405–1415. [Google Scholar] [CrossRef]
- Currie, S.M.; Findlay, E.G.; McHugh, B.J.; Mackellar, A.; Man, T.; Macmillan, D.; Wang, H.; Fitch, P.M.; Schwarze, J.; Davidson, D.J. The human cathelicidin LL-37 has antiviral activity against respiratory syncytial virus. PLoS ONE 2013, 8, e73659. [Google Scholar] [CrossRef]
- Bucak, I.H.; Ozturk, A.B.; Almis, H.; Cevik, M.O.; Tekin, M.; Konca, C.; Turgut, M.; Bulbul, M. Is there a relationship between low vitamin D and rotaviral diarrhea? Pediatr. Int. 2016, 58, 270–273. [Google Scholar] [CrossRef]
- Brice, D.C.; Toth, Z.; Diamond, G. LL-37 disrupts the Kaposi’s sarcoma-associated herpesvirus envelope and inhibits infection in oral epithelial cells. Antiviral Res. 2018, 158, 25–33. [Google Scholar] [CrossRef]
- Laplana, M.; Royo, J.L.; Fibla, J. Vitamin D Receptor polymorphisms and risk of enveloped virus infection: A. meta-analysis. Gene 2018, 678, 384–394. [Google Scholar] [CrossRef] [Green Version]
- Giraldo, D.M.; Cardona, A.; Urcuqui-Inchima, S. High-dose of vitamin D supplement isassociated with reduced susceptibility of monocyte-derived macrophages to dengue virusinfection and proinflammatory cytokine production: An exploratory study. Clin. Chim. Acta 2018, 478, 140–151. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, N.; Aguilar-Jimenez, W.; Rugeles, M.T. The Potential Protective Role of Vitamin D supplementation on HIV-1 Infection. Front. Immunol. 2019, 10, 2291. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Ran, Z.; Jiang, Q.; Hu, N.; Yu, B.; Zhu, L.; Shen, L.; Zhang, S.; Chen, L.; Chen, H.; et al. Vitamin D Alleviates Rotavirus Infection through a Microrna-155-5p Mediated Regulation of the TBK1/IRF3 Signaling Pathway In Vivo and In Vitro. Int. J. Mol. Sci. 2019, 20, 3562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Moreno, J.; Hernandez, J.C.; Urcuqui-Inchima, S. Effect of high doses of vitamin D supplementation on dengue virus replication, Toll-like receptor expression, and cytokine profiles on dendritic cells. Mol. Cell. Biochem. 2020, 464, 169–180. [Google Scholar] [CrossRef]
- Grant, W.B.; Lahore, H.; McDonnell, S.L.; Baggerly, C.A.; French, C.B.; Aliano, J.L.; Bhattoa, H.P. Evidence that Vitamin D Supplementation Could Reduce Risk of Influenza and COVID-19 Infections and Deaths. Nutrients 2020, 12, 988. [Google Scholar] [CrossRef] [Green Version]
- Laviano, E.; Sanchez Rubio, M.; Gonzalez-Nicolas, M.T.; Palacian, M.P.; Lopez, J.; Gilaberte, Y.; Calmarza, P.; Rezusta, A.; Serrablo, A. Association between preoperative levels of 25- hydroxyvitamin D and hospital-acquired infections after hepatobiliary surgery: A prospective study in a third-level hospital. PLoS ONE 2020, 15, e0230336. [Google Scholar] [CrossRef]
- Martineau, A.R.; Jolliffe, D.A.; Hooper, R.L.; Greenberg, L.; Aloia, J.F.; Bergman, P.; Dubnov-Raz, G.; Esposito, S.; Ganmaa, D.; Ginde, A.A.; et al. Vitamin D supplementation to prevent acute respiratory tract infections: Systematic review and meta-analysis of individual participant data. BMJ 2017, 356, i6583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martineau, A.R.; Jolliffe, D.A.; Greenberg, L.; Aloia, J.F.; Bergman, P.; Dubnov-Raz, G.; Esposito, S.; Ganmaa, D.; Ginde, A.A.; Goodall, E.C.; et al. Vitamin D supplementation to prevent acute respiratory infections: Individual participant data meta analysis. Health Technol. Assess 2019, 23. [Google Scholar] [CrossRef]
- Roth, D.E.; Jones, A.B.; Prosser, C.; Robinson, J.L.; Vohra, S. Vitamin D receptor polymorphisms and the risk of acute lower respiratory tract infection in early childhood. J. Infect. Dis. 2008, 197, 676–680. [Google Scholar] [CrossRef] [PubMed]
- Tavera-Mendoza, L.E.; White, J. Cell defenses and the sunshine vitamin. Sci. Am. 2007, 297, 62–72. [Google Scholar] [CrossRef]
- Norman, A. Minireview: Vitamin d receptor: New assignments for an already busy receptor. Endocrinology 2006, 147, 5542–5548. [Google Scholar] [CrossRef] [Green Version]
- Norman, A.; Bouillon, R. Vitamin D nutritional policy needs a vision for the future. Exp. Biol. Med. 2010, 235, 1034–1045. [Google Scholar] [CrossRef]
- Raab, W. Vitamin D- its bactericidal action. Chest 1946, 12, 409–415. [Google Scholar] [CrossRef]
- McCullough, P.J.; Lehrer, D.S. Vitamin D cod liver oil, sunshine and phototherapy: Safe, effective and forgotten tools for treating and curing tuberculosis infections—A comprehensive review. J. Steroid Biochem. Mol. Biol. 2018, 177, 21–29. [Google Scholar] [CrossRef]
- Meng, F.; Xu, R.; Wang, S.; Xu, Z.; Zhang, C.; Li, Y.; Yang, T.; Shi, L.; Fu, J.; Jiang, T.; et al. Human umbilical cord-derived mesenchymal stem cell therapy in patients with COVID-19: A phase 1 clinical trial. Signal Transduct. Target Ther. 2020, 5, 172. [Google Scholar] [CrossRef]
- Sahu, K.K.; Siddiqui, A.D.; Cerny, J. Mesenchymal stem cells in COVID-19: A journey from bench to bedside. Lab. Med. 2021, 52, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Xiao, K.; Hou, F.; Huang, X.; Li, B.; Qian, Z.R.; Xie, L. Mesenchymal stem cells: Current clinical progress in ARDS and COVID-19. Stem Cell Res. Ther. 2020, 11, 305. [Google Scholar] [CrossRef] [PubMed]
- Alcayaga-Miranda, F.; Cuenca, J.; Khoury, M. Antimicrobial activity of mesenchymal stem cells: Current status and new perspectives of antimicrobial peptide-based therapies. Front. Immunol. 2017, 8, 339. [Google Scholar] [CrossRef] [PubMed]
- Krasnodembskaya, A.; Song, Y.; Fang, X.; Gupta, N.; Serikov, V.; Lee, J.-W.; Matthay, M.A. Antibacterial effect of human mesenchymal stem cells is mediated in part from secretion of the antimicrobial peptide LL-37. Stem Cells 2010, 28, 2229–2238. [Google Scholar] [CrossRef] [Green Version]
- Sutton, M.T.; Fletcher, D.; Ghosh, S.K.; Weinberg, A.; van Heeckeren, R.; Kaur, S.; Sadeghi, Z.; Hijaz, A.; Reese, J.; Lazarus, H.M.; et al. Antimicrobial properties of mesenchymal stem cells: Therapeutic potential for cystic fibrosis infection, and treatment. Stem Cells Int. 2016, 2016, 5303048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Innovation Pharmaceuticals Inc. Brilacidin: First-in-Class Defensin-Mimetic Drug Candidate. Available online: https://static1.squarespace.com/static/5715352e20c647639137f992/t/5e9cebd48660e44c2754fa00/1587342453349/Brilacidin+for+COVID19+Overview+MOA%2C+PreClinical+Data%2C+Academic+Literature+-+4.20.20.pdf (accessed on 1 October 2020).


| Peptide | Sequence | Source | Inhibitory Concentration | Cell Line | Ref. |
|---|---|---|---|---|---|
| LL-37 | LGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTE | Neutrophils and epithelial cells | 100 ng/mL | Vero E6 cells | [82,83,84] |
| P9 | NGAICWGPCPTAFRQIGNCGHFKVRCCKIR | Mouse β-defensin-4 | ~5 μg/mL (IC50) 25 μg/mL (IC90) | FRhK-4 and Vero-E6 cells/BALB/c mice | [85] |
| P9R | NGAICWGPCPTAFRQIGNCGRFRVRCCRIR | Mouse β-defensin-4 | 0.9, 2.2 and 4.2 μg/mL (IC50) | Vero E6 cells | [86] |
| HD5 | ATCYCRTGRCATRESLSGVCEISGRLYRLCCR | Intestinal Paneth cells and neutrophils | 10 μg/mL | Caco-2 cells | [87] |
| HBD2 | GIGDPVTCLKSGAICHPVFCPRRYKQIGTCGLPGTKCCKKP | Surface of epithelial barriers | IC 50 of 2.4+/−0.1 μM | ACE2 HEK 293T cells | [88] |
| RTD1 | GFCRCLCRRGVCRCICTR | Rhesus macaque leukocytes | -------- | ------- | [89,90,91,92] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laneri, S.; Brancaccio, M.; Mennitti, C.; De Biasi, M.G.; Pero, M.E.; Pisanelli, G.; Scudiero, O.; Pero, R. Antimicrobial Peptides and Physical Activity: A Great Hope against COVID 19. Microorganisms 2021, 9, 1415. https://doi.org/10.3390/microorganisms9071415
Laneri S, Brancaccio M, Mennitti C, De Biasi MG, Pero ME, Pisanelli G, Scudiero O, Pero R. Antimicrobial Peptides and Physical Activity: A Great Hope against COVID 19. Microorganisms. 2021; 9(7):1415. https://doi.org/10.3390/microorganisms9071415
Chicago/Turabian StyleLaneri, Sonia, Mariarita Brancaccio, Cristina Mennitti, Margherita G. De Biasi, Maria Elena Pero, Giuseppe Pisanelli, Olga Scudiero, and Raffaela Pero. 2021. "Antimicrobial Peptides and Physical Activity: A Great Hope against COVID 19" Microorganisms 9, no. 7: 1415. https://doi.org/10.3390/microorganisms9071415
APA StyleLaneri, S., Brancaccio, M., Mennitti, C., De Biasi, M. G., Pero, M. E., Pisanelli, G., Scudiero, O., & Pero, R. (2021). Antimicrobial Peptides and Physical Activity: A Great Hope against COVID 19. Microorganisms, 9(7), 1415. https://doi.org/10.3390/microorganisms9071415