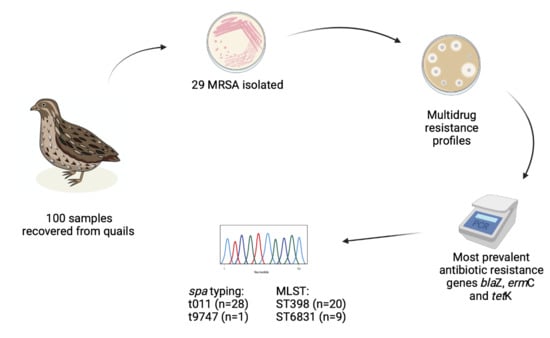
Prevalence and Characteristics of Multidrug-Resistant Livestock-Associated Methicillin-Resistant Staphylococcus aureus (LA-MRSA) CC398 Isolated from Quails (Coturnix Coturnix Japonica) Slaughtered for Human Consumption
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Bacterial Isolates
2.2. Antimicrobial Susceptibility Testing
2.3. Antimicrobial Resistance and Virulence Genes
2.4. Molecular Typing
3. Results
3.1. Antimicrobial Resistance and Virulence
3.2. Molecular Typing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Cheung, G.Y.C.; Bae, J.S.; Otto, M. Pathogenicity and virulence of Staphylococcus aureus. Virulence 2021, 12, 547–569. [Google Scholar] [CrossRef]
- Silva, V.; Capelo, J.L.; Igrejas, G.; Poeta, P. Molecular Epidemiology of Staphylococcus aureus Lineages in Wild Animals in Europe: A Review. Antibiotics 2020, 9, 122. [Google Scholar] [CrossRef]
- Cuny, C.; Wieler, L.; Witte, W. Livestock-Associated MRSA: The Impact on Humans. Antibiotics 2015, 4, 521–543. [Google Scholar] [CrossRef]
- Jayaweera, J.A.A.S.; Kumbukgolla, W.W. Antibiotic resistance patterns of methicillin-resistant Staphylococcus aureus (MRSA) isolated from livestock and associated farmers in Anuradhapura, Sri Lanka. Germs 2017, 7, 132. [Google Scholar] [CrossRef]
- Watkins, R.R.; Holubar, M.; David, M.Z. Antimicrobial Resistance in Methicillin-Resistant Staphylococcus aureus to Newer Antimicrobial Agents. Antimicrob. Agents Chemother. 2019, 63, e01216–19. [Google Scholar] [CrossRef]
- Schmithausen, R.M.; Schulze-Geisthoevel, S.V.; Heinemann, C.; Bierbaum, G.; Exner, M.; Petersen, B.; Steinhoff-Wagner, J. Reservoirs and Transmission Pathways of Resistant Indicator Bacteria in the Biotope Pig Stable and along the Food Chain: A Review from a One Health Perspective. Sustainability 2018, 10, 3967. [Google Scholar] [CrossRef]
- Suleyman, G.; Alangaden, G.; Bardossy, A.C. The Role of Environmental Contamination in the Transmission of Nosocomial Pathogens and Healthcare-Associated Infections. Curr. Infect. Dis. Rep. 2018, 20, 12. [Google Scholar] [CrossRef]
- Benjelloun Touimi, G.; Bennani, L.; Berrada, S.; Moussa, B.; Bennani, B. Prevalence and antibiotic resistance profiles of Staphylococcus sp. isolated from food, food contact surfaces and food handlers in a Moroccan hospital kitchen. Lett. Appl. Microbiol. 2020, 70, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Silva, V.; Caniça, M.; Capelo, J.L.; Igrejas, G.; Poeta, P. Diversity and genetic lineages of environmental staphylococci: A surface water overview. FEMS Microbiol. Ecol. 2020, 96, fiaa191. [Google Scholar] [CrossRef] [PubMed]
- Cuny, C.; Köck, R.; Witte, W. Livestock associated MRSA (LA-MRSA) and its relevance for humans in Germany. Int. J. Med. Microbiol. 2013, 303, 331–337. [Google Scholar] [CrossRef]
- Bernier-Lachance, J.; Arsenault, J.; Usongo, V.; Parent, É.; Labrie, J.; Jacques, M.; Malouin, F.; Archambault, M. Prevalence and characteristics of Livestock-Associated Methicillin-Resistant Staphylococcus aureus (LA-MRSA) isolated from chicken meat in the province of Quebec, Canada. PLoS ONE 2020, 15, e0227183. [Google Scholar] [CrossRef] [PubMed]
- Kinross, P.; Petersen, A.; Skov, R.; Van Hauwermeiren, E.; Pantosti, A.; Laurent, F.; Voss, A.; Kluytmans, J.; Struelens, M.J.; Heuer, O. Livestock-associated meticillin-resistant Staphylococcus aureus (MRSA) among human MRSA isolates, European Union/European Economic Area countries, 2013. Eurosurveillance 2017, 22, 16–696. [Google Scholar] [CrossRef]
- Aklilu, E.; Chia, H.Y. First mecC and mecA Positive Livestock-Associated Methicillin Resistant Staphylococcus aureus (mecC MRSA/LA-MRSA) from Dairy Cattle in Malaysia. Microorganisms 2020, 8, 147. [Google Scholar] [CrossRef]
- Tanomsridachchai, W.; Changkaew, K.; Changkwanyeun, R.; Prapasawat, W.; Intarapuk, A.; Fukushima, Y.; Yamasamit, N.; Flav Kapalamula, T.; Nakajima, C.; Suthienkul, O.; et al. Antimicrobial Resistance and Molecular Characterization of Methicillin-Resistant Staphylococcus aureus Isolated from Slaughtered Pigs and Pork in the Central Region of Thailand. Antibiotics 2021, 10, 206. [Google Scholar] [CrossRef]
- Price, L.B.; Stegger, M.; Hasman, H.; Aziz, M.; Larsen, J.; Andersen, P.S.; Pearson, T.; Waters, A.E.; Foster, J.T.; Schupp, J.; et al. Staphylococcus aureus CC398: Host adaptation and emergence of methicillin resistance in livestock. MBio 2012, 3, e00305-11. [Google Scholar] [CrossRef]
- Sousa, M.; Silva, N.; Manageiro, V.; Ramos, S.; Coelho, A.; Gonçalves, D.; Caniça, M.; Torres, C.; Igrejas, G.; Poeta, P. First report on MRSA CC398 recovered from wild boars in the north of Portugal. Are we facing a problem? Sci. Total Environ. 2017, 596–597, 26–31. [Google Scholar] [CrossRef]
- Chueahiran, S.; Yindee, J.; Boonkham, P.; Suanpairintr, N.; Chanchaithong, P. Methicillin-Resistant Staphylococcus aureus Clonal Complex 398 as a Major MRSA Lineage in Dogs and Cats in Thailand. Antibiotics 2021, 10, 243. [Google Scholar] [CrossRef] [PubMed]
- Sieber, R.N.; Larsen, A.R.; Urth, T.R.; Iversen, S.; Møller, C.H.; Skov, R.L.; Larsen, J.; Stegger, M. Genome investigations show host adaptation and transmission of LA-MRSA CC398 from pigs into Danish healthcare institutions. Sci. Rep. 2019, 9, 1–10. [Google Scholar]
- Anadón, A. WS14 The EU ban of antibiotics as feed additives (2006): Alternatives and consumer safety. J. Vet. Pharmacol. Ther. 2006, 29, 41–44. [Google Scholar] [CrossRef]
- European Medicines Agency. Sales of Veterinary Antimicrobial Agents in 31 European Countries in 2017. In ESVAC Report—Ninth ESVAC Report; European Medicines Agency: Amsterdam, The Netherlands, 2017. [Google Scholar]
- (ECDC), European Centre for Disease Prevention and Control; (EFSA), European Food Safety Authority; (EMA), European Medicines Agency. ECDC/EFSA/EMA second joint report on the integrated analysis of the consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from humans and food-producing animals: Joint Interagency Antimicrobial Consumption and Resistan. EFSA J. 2017, 15, e04872. [Google Scholar]
- Landoni, M.F.; Albarellos, G. The use of antimicrobial agents in broiler chickens. Vet. J. 2015, 205, 21–27. [Google Scholar] [CrossRef]
- Tekeli, I.O.; Turk, E.; Durna Corum, D.; Corum, O.; Kirgiz, F.C.; Uney, K. Pharmacokinetics, bioavailability and tissue residues of doxycycline in Japanese quails (Coturnix coturnix japonica) after oral administration. Food Addit. Contam. Part A 2020, 37, 2082–2092. [Google Scholar] [CrossRef]
- Santhi, D.; Kalaikannan, A. Japanese quail (Coturnix coturnix japonica) meat: Characteristics and value addition. Worlds Poult. Sci. J. 2017, 73, 337–344. [Google Scholar] [CrossRef]
- Conceição, T.; De Lencastre, H.; Aires-De-Sousa, M. Frequent isolation of methicillin resistant Staphylococcus aureus (MRSA) ST398 among healthy pigs in Portugal. PLoS ONE 2017, 12, e0175340. [Google Scholar] [CrossRef] [PubMed]
- Couto, N.; Belas, A.; Kadlec, K.; Schwarz, S. Clonal diversity, virulence patterns and antimicrobial and biocide susceptibility among human, animal and environmental MRSA in Portugal. J. Antimicrob. Chemother. 2015, 2008, 2483–2487. [Google Scholar] [CrossRef]
- Santos, V.; Gomes, A.; Ruiz-Ripa, L.; Mama, O.M.; Sabença, C.; Sousa, M.; Silva, V.; Sousa, T.; Vieira-Pinto, M.; Igrejas, G.; et al. Methicillin-Resistant Staphylococcus aureus CC398 in Purulent Lesions of Piglets and Fattening Pigs in Portugal. Microb. Drug Resist. 2020, 26, 850–856. [Google Scholar] [CrossRef]
- Silva, V.; Gabriel, S.I.; Borrego, S.B.; Tejedor-Junco, M.T.; Manageiro, V.; Ferreira, E.; Reis, L.; Caniça, M.; Capelo, J.L.; Igrejas, G.; et al. Antimicrobial Resistance and Genetic Lineages of Staphylococcus aureus from Wild Rodents: First Report of mecC-Positive Methicillin-Resistant S. aureus (MRSA) in Portugal. Animals 2021, 11, 1537. [Google Scholar] [CrossRef]
- Silva, V.; Almeida, F.; Carvalho, J.A.; Castro, A.P.; Ferreira, E.; Manageiro, V.; Tejedor-Junco, M.T.; Caniça, M.; Igrejas, G.; Poeta, P. Emergence of community-acquired methicillin-resistant Staphylococcus aureus EMRSA-15 clone as the predominant cause of diabetic foot ulcer infections in Portugal. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 179–186. [Google Scholar] [CrossRef]
- Zhang, K.; Sparling, J.; Chow, B.L.; Elsayed, S.; Hussain, Z.; Church, D.L.; Gregson, D.B.; Louie, T.; Conly, J.M. New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J. Clin. Microbiol. 2004, 42, 4947–4955. [Google Scholar] [CrossRef] [PubMed]
- Van Wamel, W.J.B.; Rooijakkers, S.H.M.; Ruyken, M.; van Kessel, K.P.M.; van Strijp, J.A.G. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J. Bacteriol. 2006, 188, 1310–1315. [Google Scholar] [CrossRef] [PubMed]
- Harmsen, D.; Claus, H.; Witte, W.; Rothgänger, J.; Claus, H.; Turnwald, D.; Vogel, U. Typing of Methicillin-Resistant Staphylococcus aureus in a University Hospital Setting by Using Novel Software for spa Repeat Determination and Database Management. J. Clin. Microbiol. 2003, 41, 5442–5448. [Google Scholar] [CrossRef]
- Enright, M.C.; Day, N.P.; Davies, C.E.; Peacock, S.J.; Spratt, B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 2000, 38, 1008–1015. [Google Scholar] [CrossRef]
- Shopsin, B.; Mathema, B.; Alcabes, P.; Said-Salim, B.; Lina, G.; Matsuka, A.; Martinez, J.; Kreiswirth, B.N. Prevalence of agr specificity groups among Staphylococcus aureus strains colonizing children and their guardians. J. Clin. Microbiol. 2003, 41, 456–459. [Google Scholar] [CrossRef]
- Zhang, K.; McClure, J.-A.; Elsayed, S.; Louie, T.; Conly, J.M. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 2005, 43, 5026–5033. [Google Scholar] [CrossRef]
- Saadati, A.; Mashak, Z.; Yarmand, M.S. Prevalence and Molecular Characterization of Enterotoxin- and Antibiotic Resistance-Encoding Genes in the Methicillin-resistant Staphylococcus aureus Recovered from Poultry Meat. Egypt. J. Vet. Sci. 2021, 52, 163–173. [Google Scholar]
- Pondit, A.; Haque, Z.F.; Sabuj, A.A.M.; Khan, M.S.R.; Saha, S. Characterization of Staphylococcus aureus isolated from chicken and quail eggshell. J. Adv. Vet. Anim. Res. 2018, 5, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Saadati, A.; Mashak, Z.; Yarmand, M.S. Prevalence of Staphylococcal Cassette Chromosome mec and Panton-Valentine Leukocidin Gene Amongst the Methicillin-resistant Staphylococcus aureus Strains Isolated from Fowl Meat. Int. J. Enteric. Pathog. 2019, 7, 93–98. [Google Scholar] [CrossRef]
- Geenen, P.L.; Graat, E.A.M.; Haenen, A.; Hengeveld, P.D.; Van Hoek, A.H.A.M.; Huijsdens, X.W.; Kappert, C.C.; Lammers, G.A.C.; Van Duijkeren, E.; Van De Giessen, A.W. Prevalence of livestock-associated MRSA on Dutch broiler farms and in people living and/or working on these farms. Epidemiol. Infect. 2013, 141, 1099–1108. [Google Scholar] [CrossRef]
- Bounar-Kechih, S.; Taha Hamdi, M.; Aggad, H.; Meguenni, N.; Cantekin, Z. Carriage Methicillin-Resistant Staphylococcus aureus in Poultry and Cattle in Northern Algeria. Vet. Med. Int. 2018, 2018, 4636121. [Google Scholar] [CrossRef] [PubMed]
- Benrabia, I.; Hamdi, T.M.; Shehata, A.A.; Neubauer, H.; Wareth, G. Methicillin-Resistant Staphylococcus aureus (MRSA) in Poultry Species in Algeria: Long-Term Study on Prevalence and Antimicrobial Resistance. Vet. Sci. 2020, 7, 54. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Larsen, J.; Kjeldgaard, J.; Andersen, P.S.; Skov, R.; Ingmer, H. Methicillin-resistant and -susceptible Staphylococcus aureus from retail meat in Denmark. Int. J. Food Microbiol. 2017, 249, 72–76. [Google Scholar] [CrossRef]
- Fox, A.; Pichon, B.; Wilkinson, H.; Doumith, M.; Hill, R.L.R.; McLauchlin, J.; Kearns, A.M. Detection and molecular characterization of Livestock-Associated MRSA in raw meat on retail sale in North West England. Lett. Appl. Microbiol. 2017, 64, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Abdalrahman, L.S.; Stanley, A.; Wells, H.; Fakhr, M.K. Isolation, Virulence, and Antimicrobial Resistance of Methicillin-Resistant Staphylococcus aureus (MRSA) and Methicillin Sensitive Staphylococcus aureus (MSSA) Strains from Oklahoma Retail Poultry Meats. Int. J. Environ. Res. Public Health 2015, 12, 6148–6161. [Google Scholar] [CrossRef] [PubMed]
- Callens, B.; Cargnel, M.; Sarrazin, S.; Dewulf, J.; Hoet, B.; Vermeersch, K.; Wattiau, P.; Welby, S. Associations between a decreased veterinary antimicrobial use and resistance in commensal Escherichia coli from Belgian livestock species (2011–2015). Prev. Vet. Med. 2018, 157, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Roth, N.; Käsbohrer, A.; Mayrhofer, S.; Zitz, U.; Hofacre, C.; Domig, K.J. The application of antibiotics in broiler production and the resulting antibiotic resistance in Escherichia coli: A global overview. Poult. Sci. 2019, 98, 1791–1804. [Google Scholar] [CrossRef]
- Petersen, A.; Stegger, M.; Heltberg, O.; Christensen, J.; Zeuthen, A.; Knudsen, L.K.; Urth, T.; Sorum, M.; Schouls, L.; Larsen, J.; et al. Epidemiology of methicillin-resistant Staphylococcus aureus carrying the novel mecC gene in Denmark corroborates a zoonotic reservoir with transmission to humans. Clin. Microbiol. Infect. 2013, 19, E16–E22. [Google Scholar] [CrossRef]
- Pesciaroli, M.; Magistrali, C.F.; Filippini, G.; Epifanio, E.M.; Lovito, C.; Marchi, L.; Maresca, C.; Massacci, F.R.; Orsini, S.; Scoccia, E.; et al. Antibiotic-resistant commensal Escherichia coli are less frequently isolated from poultry raised using non-conventional management systems than from conventional broiler. Int. J. Food Microbiol. 2020, 314, 108391. [Google Scholar] [CrossRef]
- Kadlec, K.; Feßler, A.T.; Hauschild, T.; Schwarz, S. Novel and uncommon antimicrobial resistance genes in livestock-associated methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 2012, 18, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Nhung, N.T.; Chansiripornchai, N.; Carrique-Mas, J.J. Antimicrobial Resistance in Bacterial Poultry Pathogens: A Review. Front. Vet. Sci. 2017, 4, 126. [Google Scholar] [CrossRef]
- Kadlec, K.; Ehricht, R.; Monecke, S.; Steinacker, U.; Kaspar, H.; Mankertz, J.; Schwarz, S. Diversity of antimicrobial resistance pheno- and genotypes of methicillin-resistant Staphylococcus aureus ST398 from diseased swine. J. Antimicrob. Chemother. 2009, 64, 1156–1164. [Google Scholar] [CrossRef]
- Jiménez-Belenguer, A.; Doménech, E.; Villagrá, A.; Fenollar, A.; Ferrús, M.A. Antimicrobial resistance of Escherichia coli isolated in newly-hatched chickens and effect of amoxicillin treatment during their growth. Avian Pathol. 2016, 45, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Österberg, J.; Wingstrand, A.; Nygaard Jensen, A.; Kerouanton, A.; Cibin, V.; Barco, L.; Denis, M.; Aabo, S.; Bengtsson, B. Antibiotic resistance in Escherichia coli from pigs in organic and conventional farming in four European countries. PLoS ONE 2016, 11, e0157049. [Google Scholar] [CrossRef] [PubMed]
- Dorado-García, A.; Mevius, D.J.; Jacobs, J.J.H.; Van Geijlswijk, I.M.; Mouton, J.W.; Wagenaar, J.A.; Heederik, D.J. Quantitative assessment of antimicrobial resistance in livestock during the course of a nationwide antimicrobial use reduction in the Netherlands. J. Antimicrob. Chemother. 2016, 71, 3607–3619. [Google Scholar] [CrossRef]
- Cuny, C.; Abdelbary, M.; Layer, F.; Werner, G.; Witte, W. Prevalence of the immune evasion gene cluster in Staphylococcus aureus CC398. Vet. Microbiol. 2015, 177, 219–223. [Google Scholar] [CrossRef] [PubMed]
- Argudín, M.A.; Tenhagen, B.A.; Fetsch, A.; Sachsenröder, J.; Käsbohrer, A.; Schroeter, A.; Hammer, J.A.; Hertwig, S.; Helmuth, R.; Bräunig, J.; et al. Virulence and Resistance Determinants of German Staphylococcus aureus ST398 Isolates from Nonhuman Sources. Appl. Environ. Microbiol. 2011, 77, 3052–3060. [Google Scholar] [CrossRef]
- Stegger, M.; Liu, C.M.; Larsen, J.; Soldanova, K.; Aziz, M.; Contente-Cuomo, T.; Petersen, A.; Vandendriessche, S.; Jiménez, J.N.; Mammina, C.; et al. Rapid Differentiation between Livestock-Associated and Livestock-Independent Staphylococcus aureus CC398 Clades. PLoS ONE 2013, 8, e79645. [Google Scholar] [CrossRef]
- Kraushaar, B.; Hammerl, J.A.; Kienöl, M.; Heinig, M.L.; Sperling, N.; Thanh, M.D.; Reetz, J.; Jäckel, C.; Fetsch, A.; Hertwig, S. Acquisition of virulence factors in livestock-associated MRSA: Lysogenic conversion of CC398 strains by virulence gene-containing phages. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Islam, M.Z.; Espinosa-Gongora, C.; Damborg, P.; Sieber, R.N.; Munk, R.; Husted, L.; Moodley, A.; Skov, R.; Larsen, J.; Guardabassi, L. Horses in Denmark Are a Reservoir of Diverse Clones of Methicillin-Resistant and -Susceptible Staphylococcus aureus. Front. Microbiol. 2017, 8, 543. [Google Scholar] [CrossRef] [PubMed]
- El-Adawy, H.; Ahmed, M.; Hotzel, H.; Monecke, S.; Schulz, J.; Hartung, J.; Ehricht, R.; Neubauer, H.; Hafez, H.M. Characterization of methicillin-resistant Staphylococcus aureus isolated from healthy turkeys and broilers using DNA microarrays. Front. Microbiol. 2016, 7, 2019. [Google Scholar] [CrossRef]
- Kittler, S.; Seinige, D.; Meemken, D.; Müller, A.; Wendlandt, S.; Ehricht, R.; Monecke, S.; Kehrenberg, C. Characteristics of methicillin-resistant Staphylococcus aureus from broiler farms in Germany are rather lineage- than source-specific. Poult. Sci. 2019, 98, 6903–6913. [Google Scholar] [CrossRef]
- Wendlandt, S.; Kadlec, K.; Feßler, A.T.; Monecke, S.; Ehricht, R.; van de Giessen, A.W.; Hengeveld, P.D.; Huijsdens, X.; Schwarz, S.; van Duijkeren, E. Resistance phenotypes and genotypes of methicillin-resistant Staphylococcus aureus isolates from broiler chickens at slaughter and abattoir workers. J. Antimicrob. Chemother. 2013, 68, 2458–2463. [Google Scholar] [CrossRef]
- Monecke, S.; Ruppelt, A.; Wendlandt, S.; Schwarz, S.; Slickers, P.; Ehricht, R.; de Jäckel, S.C. Genotyping of Staphylococcus aureus isolates from diseased poultry. Vet. Microbiol. 2013, 162, 806–812. [Google Scholar] [CrossRef]
- Van Meurs, M.; Schellekens, J.J.A.; De Neeling, A.J.; Duim, B.; Schneeberger, P.M.; Hermans, M.H.A. Real-time PCR to distinguish livestock-associated (ST398) from non-livestock-associated (methicillin-resistant) Staphylococcus aureus. Infection 2013, 41, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Heikinheimo, A.; Johler, S.; Karvonen, L.; Julmi, J.; Fredriksson-Ahomaa, M.; Stephan, R. New dominant spa type t2741 in livestock-associated MRSA (CC398-MRSA-V) in Finnish fattening pigs at slaughter. Antimicrob. Resist. Infect. Control 2016, 5, 6. [Google Scholar] [CrossRef] [PubMed]
- Back, S.H.; Eom, H.S.; Lee, H.H.; Lee, G.Y.; Park, K.T.; Yang, S.-J. Livestock-associated methicillin-resistant Staphylococcus aureus in Korea: Antimicrobial resistance and molecular characteristics of LA-MRSA strains isolated from pigs, pig farmers, and farm environment. J. Vet. Sci. 2020, 21, e2. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, V.; Vieira-Pinto, M.; Saraiva, C.; Manageiro, V.; Reis, L.; Ferreira, E.; Caniça, M.; Capelo, J.L.; Igrejas, G.; Poeta, P. Prevalence and Characteristics of Multidrug-Resistant Livestock-Associated Methicillin-Resistant Staphylococcus aureus (LA-MRSA) CC398 Isolated from Quails (Coturnix Coturnix Japonica) Slaughtered for Human Consumption. Animals 2021, 11, 2038. https://doi.org/10.3390/ani11072038
Silva V, Vieira-Pinto M, Saraiva C, Manageiro V, Reis L, Ferreira E, Caniça M, Capelo JL, Igrejas G, Poeta P. Prevalence and Characteristics of Multidrug-Resistant Livestock-Associated Methicillin-Resistant Staphylococcus aureus (LA-MRSA) CC398 Isolated from Quails (Coturnix Coturnix Japonica) Slaughtered for Human Consumption. Animals. 2021; 11(7):2038. https://doi.org/10.3390/ani11072038
Chicago/Turabian StyleSilva, Vanessa, Madalena Vieira-Pinto, Cândido Saraiva, Vera Manageiro, Lígia Reis, Eugénia Ferreira, Manuela Caniça, José L. Capelo, Gilberto Igrejas, and Patrícia Poeta. 2021. "Prevalence and Characteristics of Multidrug-Resistant Livestock-Associated Methicillin-Resistant Staphylococcus aureus (LA-MRSA) CC398 Isolated from Quails (Coturnix Coturnix Japonica) Slaughtered for Human Consumption" Animals 11, no. 7: 2038. https://doi.org/10.3390/ani11072038
APA StyleSilva, V., Vieira-Pinto, M., Saraiva, C., Manageiro, V., Reis, L., Ferreira, E., Caniça, M., Capelo, J. L., Igrejas, G., & Poeta, P. (2021). Prevalence and Characteristics of Multidrug-Resistant Livestock-Associated Methicillin-Resistant Staphylococcus aureus (LA-MRSA) CC398 Isolated from Quails (Coturnix Coturnix Japonica) Slaughtered for Human Consumption. Animals, 11(7), 2038. https://doi.org/10.3390/ani11072038