Boundary Matching and Interior Connectivity-Based Cluster Validity Anlysis
Abstract
:1. Introduction
2. Related Work
2.1. Typical Clustering Algorithms
2.1.1. C-Means Algorithm
| Algorithm 1. C-means Clustering Algorithm |
| Input: the number of clusters C and a dataset X containing n points. |
| Output: a set of C clusters that contains all n objects. |
| Steps: |
| 1. Initialize cluster centers by selecting C points arbitrarily. |
| 2. Repeat; |
| 3. Assign each point to one certain cluster according to the nearest neighborhood principle and a chosen measure; |
| 4. Update cluster centers by for i = 1 − C; |
| 5. Stop if a convergence criterion is satisfied; |
| 6. Otherwise, go to Step 2. |
2.1.2. DBSCAN Algorithm
2.1.3. DPC Algorithm
2.2. Typical Cluster Validity Index
2.2.1. Davies–Bouldin (DB) Index
2.2.2. Dual-Center (DC) Index
2.2.3. Gap Statistic (GS) Index
3. Materials and Methods
3.1. Boundary Matching and Connectivity
3.2. Clustering Evaluation Based on Boundary and Interior Points
- (1)
- Complementarity. The mathematical curvature and difference can reflect the varied tendency of a curve in (13), and thereby the real number of clusters c* can be found. When c < c*, R1(c) is nearly equivalent to R1(c*) since the set of boundary points is approximately unchangeable, but R2(c) < R2(c*), since the number of center points successively increase. In sum, F(c) < F(c*). Inversely, when c > c*, R1(c) successively decreases, and R2(c) tends to be flat; therefore, F(c) < F(c*). Consequently, (13) can attain a maximum when c* appears.
- (2)
- Monotonicity. Assume that c* is the real number of clusters, and when c takes its two values c1 and c2 satisfying c1 < c2 < c*, F(c1) < F(c2) < F(c*); otherwise, when c* < c1 < c2, F(c*) > F(c1) > F(c2); Hence, F(c) consists of two monotone functions at the two sides of c*, respectively. Therefore, for arbitrary two values c1 and c2 satisfying c1 < c2, c2 may refer to a more optimal clustering result. Usually, a larger value of F(c*) indicates a better clustering result.
- (3)
- Generalization. Equation (13) can provide a wide entry for any clustering algorithm only if the clustering results are available and the corresponding numbers of clusters are taken as the variable of F(c). Especially, a group of clustering results may result from different clustering algorithms and parameters, since any two clustering results are comparable according to the above monotonicity. For example, one clustering result with c1 results from C-means, and others from DBSCAN, and so on. Equation (13) can evaluate the results of any clustering algorithm and parameter in a trial-and-error way. In comparison, the existing validity indices can mainly work for a specific algorithm and parameter of the number of clusters since the center in them has to be defined, especially for the C-means algorithm.
| Algorithm 2. Evaluating Process Based on CVIBI |
| Input: a dataset X RD containing n points and clustering results from any clustering algorithm at c = 1, 2, …, cmax. |
| Output: the suggested number of clusters. |
| Steps: |
| 1. Calculate the density for each point in X according to (10); |
| 2. Partition X into boundary and interior points; |
| 3. Input clustering results at c = 1, 2, …, cmax; |
| 4. Partition each cluster into boundary and interior points; |
| 5. Compute values of bou (c) or con(c) at c equals 1, 2, …., cmax; |
| 6. Solve the optimal value of (13); |
| 7. Suggest an optimal number of clusters. |
| 8. Stop. |
4. Results and Discussion
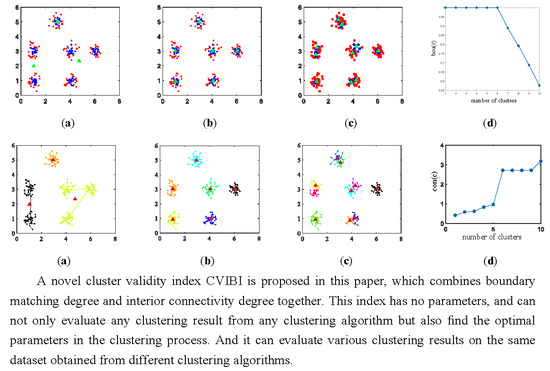
4.1. Tests on Synthetic Datasets
4.1.1. Relationship between CVIBI and the Number of Clusters
4.1.2. Relationship between CVIBI and in DBSCAN
4.1.3. CVIBI Evaluation under Various Clustering Algorithms
4.1.4. Comparison between CVIBI and Existing Validity Indices
- (1)
- Size and distribution. Groups 2 and 4 denote clusters with different sizes and distributions, and there are no overlapped clusters. The evaluation results show that except set 10, all validity indices are capable of determining the correct number of clusters no matter which clustering algorithm is used.
- (2)
- Density and shape. Groups 1 and 3 contain clusters with different densities and shapes. When the densities among clusters are so skewed as in sets 1 and 3, the three validity indices cannot find the correct number of clusters. Compared with DC and DB, the cluster numbers calculated by CVIBI are closer to the real numbers of clusters.
- (3)
- Overlap. Group 5 contains datasets with overlapped clusters. DPC is good at clustering such datasets, and the evaluation results of CVIBI are nearest to the real number of clusters. However, DC and GS with DPC give a relatively smaller number. Note the two indices are originally designed to evaluate the results for nonoverlapping clusters. Thus any two overlapped clusters may be incorrectly regarded as one cluster. For instance, the clusters in set 14 are most overlapped, and its evaluation results are the weakest. How to find the correct number of clusters in a dataset containing overlapped clusters is a difficult task for most existing validity indices, such as DB and GS; fortunately, CVIBI with DPC is effective for dealing with such a problem. In summary, DC shows performances better than CVIBI in the case of datasets containing spherical clusters.
4.2. Tests on CT Images
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Wang, T.; Wang, X.; Zhao, Z.; He, Z.; Xia, T. Measurement data classification optimization based on a novel evolutionary kernel clustering algorithm for multi-target tracking. IEEE Sens. J. 2018, 18, 3722–3733. [Google Scholar] [CrossRef]
- Nayak, P.; Vathasavai, B. Energy efficient clustering algorithm for multi-hop wireless sensor network using type-2 fuzzy logic. IEEE Sens. J. 2017, 17, 4492–4499. [Google Scholar] [CrossRef]
- Dhanachandra, N.; Chanu, Y.J. A new image segmentation method using clustering and region merging techniques. In Applications of Artificial Intelligence Techniques in Engineering; Springer: Singapore, 2019; pp. 603–614. [Google Scholar]
- Masud, M.A.; Huang, J.Z.; Wei, C.; Wang, J.; Khan, I. I-nice: A new approach for identifying the number of clusters and initial cluster centres. Inf. Sci. 2018, 466, 129–151. [Google Scholar] [CrossRef]
- Wang, J.-S.; Chiang, J.-C. A cluster validity measure with a hybrid parameter search method for the support vector clustering algorithm. Pattern Recognit. 2008, 41, 506–520. [Google Scholar] [CrossRef]
- Davies, D.L.; Bouldin, D.W. A cluster separation measure. IEEE Trans. Pattern Anal. Mach. Intell. 1979, 224–227. [Google Scholar] [CrossRef]
- Tibshirani, R.; Walther, G.; Hastie, T. Estimating the number of clusters in a data set via the gap statistic. J. R. Stat. Soc. Ser. B (Stat. Methodol.) 2001, 63, 411–423. [Google Scholar] [CrossRef]
- Xie, X.L.; Beni, G. A validity measure for fuzzy clustering. IEEE Trans. Pattern Anal. Mach. Intell. 1991, 13, 841–847. [Google Scholar] [CrossRef]
- Mehrjou, A.; Hosseini, R.; Araabi, B.N. Improved bayesian information criterion for mixture model selection. Pattern Recognit. Lett. 2016, 69, 22–27. [Google Scholar] [CrossRef]
- Dasgupta, A.; Raftery, A.E. Detecting features in spatial point processes with clutter via model-based clustering. J. Am. Stat. Assoc. 1998, 93, 294–302. [Google Scholar] [CrossRef]
- Teklehaymanot, F.K.; Muma, M.; Zoubir, A.M. Bayesian cluster enumeration criterion for unsupervised learning. IEEE Trans. Signal Process. 2018, 66, 5392–5406. [Google Scholar] [CrossRef] [Green Version]
- Arbelaitz, O.; Gurrutxaga, I.; Muguerza, J.; PéRez, J.M.; Perona, I. An extensive comparative study of cluster validity indices. Pattern Recognit. 2013, 46, 243–256. [Google Scholar] [CrossRef]
- Wang, Y.; Yue, S.; Hao, Z.; Ding, M.; Li, J. An unsupervised and robust validity index for clustering analysis. Soft Comput. 2019, 23, 10303–10319. [Google Scholar] [CrossRef]
- Salloum, S.; Huang, J.Z.; He, Y.; Chen, X. An asymptotic ensemble learning framework for big data analysis. IEEE Access 2018, 7, 3675–3693. [Google Scholar] [CrossRef]
- Chen, X.; Hong, W.; Nie, F.; He, D.; Yang, M. Spectral clustering of large-scale data by directly solving normalized cut. In Proceedings of the 24th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, London, UK, 19–23 August 2018. [Google Scholar]
- Havens, T.C.; Bezdek, J.C.; Leckie, C.; Hall, L.O.; Palaniswami, M. Fuzzy c-means algorithms for very large data. IEEE Trans. Fuzzy Syst. 2012, 20, 1130–1146. [Google Scholar] [CrossRef]
- Dhanachandra, N.; Manglem, K.; Chanu, Y.J. Image segmentation using K-means clustering algorithm and subtractive clustering algorithm. Procedia Comput. Sci. 2015, 54, 764–771. [Google Scholar] [CrossRef] [Green Version]
- Bagirov, A.M.; Ugon, J.; Webb, D. Fast modified global k-means algorithm for incremental cluster construction. Pattern Recognit. 2011, 44, 866–876. [Google Scholar] [CrossRef]
- Cai, W.; Chen, S.; Zhang, D. Fast and robust fuzzy c-means clustering algorithms incorporating local information for image segmentation. Pattern Recognit. 2007, 40, 825–838. [Google Scholar] [CrossRef] [Green Version]
- Wu, K.-L.; Yang, M.-S. A cluster validity index for fuzzy clustering. Pattern Recognit. Lett. 2005, 26, 1275–1291. [Google Scholar] [CrossRef]
- Ester, M.; Kriegel, H.-P.; Sander, J.; Xu, X. A density-based algorithm for discovering clusters in large spatial databases with noise. KDD 1996, 96, 226–231. [Google Scholar]
- Ma, E.W.; Chow, T.W. A new shifting grid clustering algorithm. Pattern Recognit. 2004, 37, 503–514. [Google Scholar] [CrossRef]
- Rodriguez, A.; Laio, A. Clustering by fast search and find of density peaks. Science 2014, 344, 1492–1496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, M.; Ding, S.; Xue, Y. A novel density peaks clustering algorithm for mixed data. Pattern Recognit. Lett. 2017, 97, 46–53. [Google Scholar] [CrossRef]
- Ding, S.; Du, M.; Sun, T.; Xu, X.; Xue, Y. An entropy-based density peaks clustering algorithm for mixed type data employing fuzzy neighborhood. Knowl. Based Syst. 2017, 133, 294–313. [Google Scholar] [CrossRef]
- Du, M.; Ding, S.; Jia, H. Study on density peaks clustering based on k-nearest neighbors and principal component analysis. Knowl. Based Syst. 2016, 99, 135–145. [Google Scholar] [CrossRef]
- Krammer, P.; Habala, O.; Hluchý, L. Transformation regression technique for data mining. In Proceedings of the 2016 IEEE 20th Jubilee International Conference on Intelligent Engineering Systems (INES), Budapest, Hungary, 30 June–2 July 2016. [Google Scholar]
- Yan, X.; Zhang, C.; Zhang, S. Toward databases mining: Pre-processing collected data. Appl. Artif. Intell. 2003, 17, 545–561. [Google Scholar] [CrossRef]
- Ankerst, M.; Breunig, M.M.; Kriegel, H.-P.; Sander, J. OPTICS: Ordering points to identify the clustering structure. ACM Sigmod Rec. 1999, 28, 49–60. [Google Scholar] [CrossRef]
- Hung, W.-L.; Yang, M.-S. Similarity measures of intuitionistic fuzzy sets based on Hausdorff distance. Pattern Recognit. Lett. 2004, 25, 1603–1611. [Google Scholar] [CrossRef]
- Qian, R.; Wei, Y.; Shi, H.; Li, J.; Liu, J. Weakly supervised scene parsing with point-based distance metric learning. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019. [Google Scholar]
- Bandaru, S.; Ng, A.H.; Deb, K. Data mining methods for knowledge discovery in multi-objective optimization: Part A-Survey. Expert Syst. Appl. 2017, 70, 139–159. [Google Scholar] [CrossRef] [Green Version]
- Yue, S.; Wang, J.; Wang, J.; Bao, X. A new validity index for evaluating the clustering results by partitional clustering algorithms. Soft Comput. 2016, 20, 1127–1138. [Google Scholar] [CrossRef]
- Khan, M.M.R.; Arif, R.B.; Siddique, M.A.B.; Oishe, M.R. Study and observation of the variation of accuracies of KNN, SVM, LMNN, ENN algorithms on eleven different datasets from UCI machine learning repository. In Proceedings of the 2018 4th International Conference on Electrical Engineering and Information & Communication Technology (iCEEiCT), Dhaka, Bangladesh, 13–15 September 2018. [Google Scholar]
- Nie, F.; Wang, X.; Huang, H. Clustering and projected clustering with adaptive neighbors. In Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 24–27 August 2014. [Google Scholar]
- Hubert, L.J. Some applications of graph theory to clustering. Psychometrika 1974, 39, 283–309. [Google Scholar] [CrossRef]
- Yin, M.; Xie, S.; Wu, Z.; Zhang, Y.; Gao, J. Subspace clustering via learning an adaptive low-rank graph. IEEE Trans. Image Process. 2018, 27, 3716–3728. [Google Scholar] [CrossRef] [PubMed]


















| Types/Algorithm | C-Means | DBSCAN | DPC |
|---|---|---|---|
| Arbitrary shape | × | √ | √/× |
| Density-diversity | √/× | × | √/× |
| overlap | √/× | × | √ |
| High-dimension | √ | × | √/× |
| The number of clusters | × | × | × |
| Name | Clusters | Dimension | Number of Objects | Number of Each Cluster |
|---|---|---|---|---|
| Set 1 | 3 | 2 | 600 | 83/164/353 |
| Set 2 | 4 | 2 | 350 | 30/60/120/240 |
| Set 3 | 5 | 2 | 830 | 30/60/120/240/480 |
| Set 4 | 3 | 2 | 420 | 60/120/240 |
| Set 5 | 4 | 2 | 900 | 60/120/240/480 |
| Set 6 | 5 | 2 | 1860 | 60/120/240/480/960 |
| Set 7 | 3 | 2 | 1018 | 341/336/341 |
| Set 8 | 4 | 2 | 404 | 134/90/90/90 |
| Set 9 | 5 | 2 | 494 | 134/90/90/90/90 |
| Set 10 | 3 | 2 | 360 | 120/120/120 |
| Set 11 | 4 | 2 | 800 | 200/200/200/200 |
| Set 12 | 5 | 2 | 1000 | 200/200/200/200/200 |
| Set 13 | 3 | 2 | 600 | 200/200/200 |
| Set 14 | 4 | 2 | 400 | 100/100/100/100 |
| Set 15 | 5 | 2 | 1000 | 200/200/200/200/200 |
| Dataset and Algorithm | DB | DC | GS | CVIBI | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C-Means | DBSCAN | DPC | C-Means | DBSCAN | DPC | C-Means | DBSCAN | DPC | C-Means | DBSCAN | DPC | |
| Set 1 | 3√ | 2 | 3√ | 3√ | 2 | 3√ | 3√ | 2 | 3√ | 3√ | 3√ | 3√ |
| Set 2 | 4√ | 3 | 4√ | 4√ | 3 | 4√ | 4√ | 2 | 4√ | 4√ | 4√ | 4√ |
| Set 3 | 4 | 6 | 4 | 4 | 4 | 5√ | 3 | 3 | 5√ | 5√ | 5√ | 5√ |
| Set 4 | 3√ | 3√ | 3√ | 3√ | 3√ | 3√ | 3√ | 3√ | 3√ | 3√ | 3√ | 3√ |
| Set 5 | 4√ | 4√ | 4√ | 4√ | 4√ | 4√ | 4√ | 4√ | 4√ | 4√ | 4√ | 4√ |
| Set 6 | 4 | 5√ | 4 | 5√ | 4 | 5√ | 4 | 5√ | 5√ | 5√ | 5√ | 5√ |
| Set 7 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3√ | 3√ |
| Set 8 | 2 | 4√ | 2 | 4√ | 2 | 4√ | 2 | 2 | 4√ | 2 | 4√ | 4√ |
| Set 9 | 2 | 5√ | 2 | 5√ | 2 | 5√ | 2 | 2 | 5√ | 2 | 5√ | 5√ |
| Set 10 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3√ |
| Set 11 | 4√ | 4√ | 4√ | 4√ | 4√ | 4√ | 4√ | 4√ | 4√ | 4√ | 4√ | 4√ |
| Set 12 | 5√ | 5√ | 5√ | 5√ | 5√ | 5√ | 5√ | 5√ | 5√ | 5√ | 5√ | 5√ |
| Set 13 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 2 | 1 | 2 | 1 | 2 |
| Set 14 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 2 |
| Set 15 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 2 | 1 | 2 | 1 | 4 |
| Dataset and Algorithm | DB | DC | GS | CVIBI | ||||
|---|---|---|---|---|---|---|---|---|
| DPC | C-Means | DPC | C-Means | DPC | C-Means | DPC | C-Means | |
| Set 1 | 4 | 5√ | 3√ | 5√ | 3√ | 5√ | 3√ | 5√ |
| Set 2 | 2 | 3 | 7√ | 3 | 7√ | 5√ | 7√ | 5√ |
| Set 3 | 2 | 4 | 5√ | 4 | 5√ | 5√ | 5√ | 5√ |
| Set 4 | 2 | 5√ | 3√ | 5√ | 3√ | 5√ | 3√ | 5√ |
| Set 5 | 4√ | 5√ | 4√ | 5√ | 4√ | 5√ | 4√ | 5√ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Yue, S.; Wang, Y.; Ding, M.; Li, J.; Wang, Z. Boundary Matching and Interior Connectivity-Based Cluster Validity Anlysis. Appl. Sci. 2020, 10, 1337. https://doi.org/10.3390/app10041337
Li Q, Yue S, Wang Y, Ding M, Li J, Wang Z. Boundary Matching and Interior Connectivity-Based Cluster Validity Anlysis. Applied Sciences. 2020; 10(4):1337. https://doi.org/10.3390/app10041337
Chicago/Turabian StyleLi, Qi, Shihong Yue, Yaru Wang, Mingliang Ding, Jia Li, and Zeying Wang. 2020. "Boundary Matching and Interior Connectivity-Based Cluster Validity Anlysis" Applied Sciences 10, no. 4: 1337. https://doi.org/10.3390/app10041337
APA StyleLi, Q., Yue, S., Wang, Y., Ding, M., Li, J., & Wang, Z. (2020). Boundary Matching and Interior Connectivity-Based Cluster Validity Anlysis. Applied Sciences, 10(4), 1337. https://doi.org/10.3390/app10041337