A Robust Brain MRI Segmentation and Bias Field Correction Method Integrating Local Contextual Information into a Clustering Model
Abstract
:Featured Application
Abstract
1. Introduction
2. Related Work
2.1. Bias-Corrected Fuzzy C-Means (BCFCM)
2.2. Fuzzy Local Information C-Means (FLICM)
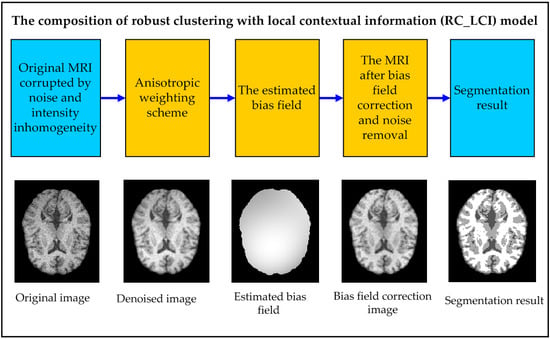
3. Robust Clustering with Local Contextual Information (RC_LCI)
3.1. Anisotropic Weighting Scheme
3.2. Bias Field Framework
3.3. Energy Formulation
3.4. Energy Minimization
| Algorithm 1. RC_LCI segmentation algorithm. |
| 1. Input: Brain MRI to be segmented. 2. Image = denoising(Image, ) % Update each pixel according to Equations (8)–(11). 3. Basis = GetBasisOrder3(size(Image)) % Obtain basis functions according to Equation (14). 4. Randomly initialize bias field b, cluster center C, and membership function M. 5. while C(n) - C(n-1) > 0.001 do 6. for i = 1 : size(Basis, 3) 7. v(i) = Basis ∗ Image ∗ C ∗ M 8. for j = i : size(Basis, 3) 9. A(i, j) = Basis ∗ C2 ∗ M 10. A(j, i) = A(i, j) 11. end for 12. end for 13. w = inv(A) ∗ v 14. for i = 1 : size(Basis, 3) 15. b = b + w(k) ∗ Basis(k) 16. end for 17. for n = 1 : size(M, 3) 18. N = b ∗ Image ∗ M 19. D = b2 ∗ M 20. C(n) = N / D 21. e (i) = (Image – C(i) ∗ b)2 22. end for 23. N_min = min(e, [[], 3) 24. for k = 1 : size(e, 3) 25. M(:, :, k) = (N_min == k) 26. end for 27. end while 28. New_Image = C ∗ M 29. Image_bc = New_Image ./ b % Bias field correction image 30. Output: Bias field b, bias field correction image, and segmentation result. |
4. Experimental Results
4.1. Robustness to Noise
4.2. Capability of Estimating the Bias Field
4.3. Effectiveness of RC_LCI
5. Conclusions
Data Availability
Author Contributions
Funding
Conflicts of Interest
References
- González-Villà, S.; Oliver, A.; Valverde, S.; Wang, L.; Zwiggelaar, R.; Lladóa, X. A review on brain structures segmentation in magnetic resonance imaging. Artif. Intell. Med. 2016, 73, 45–69. [Google Scholar] [CrossRef]
- Dora, L.; Agrawal, S.; Panda, R.; Abraham, A. State-of-the-Art Methods for Brain Tissue Segmentation: A Review. IEEE Rev. Biomed. Eng. 2017, 10, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Zhang, Z. A Modified Robust FCM Model with Spatial Constraints for Brain MR Image Segmentation. Information 2019, 10, 74. [Google Scholar] [CrossRef]
- Song, J.; Cong, W.; Li, J. A Robust Fuzzy c-Means Clustering Model with Spatial Constraint for Brain Magnetic Resonance Image Segmentation. J. Med. Imaging Health Inform. 2018, 8, 811–816. [Google Scholar] [CrossRef]
- Jiang, X.; Wang, Q.; He, B.; Chen, S.; Li, B. Robust level set image segmentation algorithm using local correntropy-based fuzzy c-means clustering with spatial constraints. Neurocomputing 2016, 207, 22–35. [Google Scholar] [CrossRef]
- Zhao, F.; Liu, H.; Fan, J.; Chen, C.; Lan, R.; Li, N. Intuitionistic fuzzy set approach to multi-objective evolutionary clustering with multiple spatial information for image segmentation. Neurocomputing 2018, 312, 296–309. [Google Scholar] [CrossRef]
- Li, B.; Qin, J.; Wang, R.; Wang, M.; Li, X. Selective Level Set Segmentation Using Fuzzy Region Competition. IEEE Access 2016, 4, 4777–4788. [Google Scholar] [CrossRef]
- Zhang, Z.; Song, J. An Adaptive Fuzzy Level Set Model with Local Spatial Information for Medical Image Segmentation and Bias Correction. IEEE Access 2019, 7, 27322–27338. [Google Scholar] [CrossRef]
- Huang, G.; Ji, H.; Zhang, W. A fast level set method for inhomogeneous image segmentation with adaptive scale parameter. Magn. Reson. Imaging 2018, 52, 33–45. [Google Scholar] [CrossRef]
- Assaf, H.; Christopher, F.; Guilherme, M.; Elhamy, H.; Claude, B.; Sandy, N.; Daniel, L. Adaptive local window for level set segmentation of CT and MRI liver lesions. Med. Image Anal. 2017, 37, 46–55. [Google Scholar]
- Mandal, D.; Chatterjee, A.; Maitra, M. Robust medical image segmentation using particle swarm optimization aided level set based global fitting energy active contour approach. Eng. Appl. Artif. Intell. 2014, 35, 199–214. [Google Scholar] [CrossRef]
- Ali, S.; Madabhushi, A. An Integrated Region-, Boundary-, Shape-Based Active Contour for Multiple Object Overlap Resolution in Histological Imagery. IEEE Trans. Med. Imaging 2012, 31, 1448–1460. [Google Scholar] [CrossRef] [PubMed]
- Katiyar, P.; Divine, M.; Kohlhofer, U.; Quintanilla-Martinez, L.; Scholkopf, B.; Pichler, B.; Disselhorst, J. A novel unsupervised segmentation approach quantifies tumor tissue populations using multiparametric MRI: First results with histological valida-tion. Mol. Imaging Biol. 2017, 19, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Turajlic, E.; Begović, A.; Škaljo, N. Application of Artificial Neural Network for Image Noise Level Estimation in the SVD domain. Electronics 2019, 8, 163. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Z.; Si, L.; Zhang, L.; Tan, C.; Xu, J. A Non-Reference Image Denoising Method for Infrared Thermal Image Based on Enhanced Dual-Tree Complex Wavelet Optimized by Fruit Fly Algorithm and Bilateral Filter. Appl. Sci. 2017, 7, 1190. [Google Scholar] [CrossRef]
- Lu, Z.; Qiu, Y.; Zhan, T. Neutrosophic C-means clustering with local information and noise distance-based kernel metric image segmentation. J. Vis. Commun. Image R. 2019, 58, 269–276. [Google Scholar] [CrossRef]
- Mahata, N.; Kahali, S.; Adhikari, S.; Sing, J. Local contextual information and Gaussian function induced fuzzy clustering algorithm for brain MR image segmentation and intensity inhomogeneity estimation. Appl. Soft. Comput. 2018, 68, 586–596. [Google Scholar] [CrossRef]
- Song, J.; Zhang, Z. Brain Tissue Segmentation and Bias Field Correction of MR Image Based on Spatially Coherent FCM with Nonlocal Constraints. Comput. Math. Method Med. 2019, 2019, 1–13. [Google Scholar] [CrossRef]
- Pham, T.; Siarry, P.; Oulhad, H. Integrating fuzzy entropy clustering with an improved PSO for MRI brain image segmentation. Appl. Soft. Comput. 2018, 65, 230–242. [Google Scholar] [CrossRef]
- Choy, S.; Lam, S.; Yu, K.; Lee, W.; Leung, K. Fuzzy model-based clustering and its application in image segmentation. Pattern Recognit. 2017, 68, 141–157. [Google Scholar] [CrossRef]
- Singh, C.; Bala, A. A local Zernike moment-based unbiased nonlocal means fuzzy C-Means algorithm for segmentation of brain magnetic resonance images. Expert Syst. Appl. 2019, 118, 625–639. [Google Scholar] [CrossRef]
- Ahmed, M.; Yamany, S.; Mohamed, N.; Farag, A.; Moriarty, T. A modified fuzzy c-means algorithm for bias field estimation and segmentation of MRI data. IEEE Trans. Med. Imaging 2002, 21, 193–199. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, D. Robust Image Segmentation Using FCM With Spatial Constraints Based on New Kernel-Induced Distance Measure. IEEE Trans. Syst. Man Cybern. Part B-Cybern. 2004, 34, 1907–1916. [Google Scholar] [CrossRef]
- Krinidis, S.; Chatzis, V. A Robust Fuzzy Local Information C-Means Clustering Algorithm. IEEE Trans. Image Process. 2010, 19, 1328–1337. [Google Scholar] [CrossRef] [PubMed]
- Ji, Z.; Xia, Y.; Chen, Q.; Sun, Q.; Xia, D.; Feng, D. Fuzzy c-means clustering with weighted image patch for image segmentation. Appl. Soft. Comput. 2012, 12, 1659–1667. [Google Scholar] [CrossRef]
- Li, C.; Huang, R.; Ding, Z.; Gatenby, J.; Metaxas, D.; Gore, J. A Level Set Method for Image Segmentation in the Presence of Intensity Inhomogeneities with Application to MRI. IEEE Trans. Image Process. 2011, 20, 2007–2016. [Google Scholar] [PubMed]
- Li, C.; Gore, J.; Davatzikos, C. Multiplicative intrinsic component optimization (MICO) for MRI bias field estimation and tissue segmentation. Magn. Reson. Imaging 2014, 32, 913–923. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Zhao, D.; Huang, M. Image segmentation using CUDA accelerated non-local means denoising and bias correction embedded fuzzy c-means (BCEFCM). Signal Process. 2015, 122, 164–189. [Google Scholar] [CrossRef]
- BrainWeb: Simulated Brain Database. Available online: http://brainweb.bic.mni.mcgill.ca/brainweb/ (accessed on 17 August 2004).







| 67th Slice | 96th Slice | |||
|---|---|---|---|---|
| Model | Iteration | Time (s) | Iteration | Time (s) |
| MICO | 9 | 4.79 | 8 | 3.97 |
| BCFCM | 26 | 25.77 | 23 | 22.56 |
| FLICM | 44 | 6.61 | 40 | 5.83 |
| RC_LCI | 11 | 3.83 | 9 | 3.16 |
| Tissues | LIC | MICO | RC_LCI |
|---|---|---|---|
| GM | 0.768 | 0.735 | 0.842 |
| WM | 0.815 | 0.783 | 0.889 |
| CSF | 0.602 | 0.691 | 0.803 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Song, J. A Robust Brain MRI Segmentation and Bias Field Correction Method Integrating Local Contextual Information into a Clustering Model. Appl. Sci. 2019, 9, 1332. https://doi.org/10.3390/app9071332
Zhang Z, Song J. A Robust Brain MRI Segmentation and Bias Field Correction Method Integrating Local Contextual Information into a Clustering Model. Applied Sciences. 2019; 9(7):1332. https://doi.org/10.3390/app9071332
Chicago/Turabian StyleZhang, Zhe, and Jianhua Song. 2019. "A Robust Brain MRI Segmentation and Bias Field Correction Method Integrating Local Contextual Information into a Clustering Model" Applied Sciences 9, no. 7: 1332. https://doi.org/10.3390/app9071332
APA StyleZhang, Z., & Song, J. (2019). A Robust Brain MRI Segmentation and Bias Field Correction Method Integrating Local Contextual Information into a Clustering Model. Applied Sciences, 9(7), 1332. https://doi.org/10.3390/app9071332