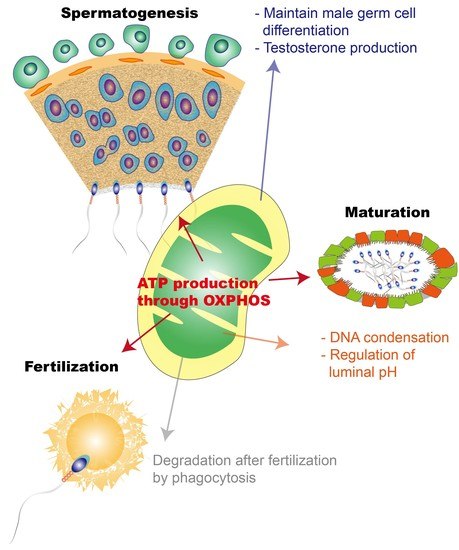
Mitochondrial Functionality in Male Fertility: From Spermatogenesis to Fertilization
Abstract
:1. Introduction
2. Mitochondria Dynamics in the Testis
2.1. Metabolic and Morphological Changes in Mitochondria during Spermatogenesis
2.2. Energy Metabolism in Germ Cell Response to Testicular Somatic Cells Depending on the Sperm Developmental State
3. Orchestrating Mitochondria in the Epididymis
3.1. Epididymal Epithelial Cells Work in a Concerted Manner to Establish Optimal Conditions for Sperm Maturation
3.2. Regulation of ROS in the Epididymis for Sperm Maturation
4. Mitochondria Is a Key Organelle for Sperm Fertility
4.1. Mitochondrial Activity of Spermatozoa in Female Reproductive Tract
4.2. Mitochondrial Proteins Associated with Male Fertility
4.3. Elimination of Paternal Mitochondrial DNA during Fertilization
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Tzameli, I. The evolving role of mitochondria in metabolism. Trends Endocrinol. Metab. 2012, 23, 417–419. [Google Scholar] [CrossRef]
- Harris, D.A.; Das, A.M. Control of mitochondrial ATP synthesis in the heart. Biochem. J. 1991, 280 Pt 3, 561–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilkerson, R.W.; Selker, J.M.; Capaldi, R.A. The cristal membrane of mitochondria is the principal site of oxidative phosphorylation. FEBS Lett. 2003, 546, 355–358. [Google Scholar] [CrossRef] [Green Version]
- Scorrano, L.; Ashiya, M.; Buttle, K.; Weiler, S.; Oakes, S.A.; Mannella, C.A.; Korsmeyer, S.J. A distinct pathway remodels mitochondrial cristae and mobilizes cytochrome c during apoptosis. Dev. Cell 2002, 2, 55–67. [Google Scholar] [CrossRef] [Green Version]
- Schagger, H.; Pfeiffer, K. The ratio of oxidative phosphorylation complexes I-V in bovine heart mitochondria and the composition of respiratory chain supercomplexes. J. Biol. Chem. 2001, 276, 37861–37867. [Google Scholar] [CrossRef]
- Acin-Perez, R.; Fernandez-Silva, P.; Peleato, M.L.; Perez-Martos, A.; Enriquez, J.A. Respiratory active mitochondrial supercomplexes. Mol. Cell 2008, 32, 529–539. [Google Scholar] [CrossRef]
- van der Bliek, A.M.; Sedensky, M.M.; Morgan, P.G. Cell Biology of the Mitochondrion. Genetics 2017, 207, 843–871. [Google Scholar] [CrossRef] [Green Version]
- Starkov, A.A. The role of mitochondria in reactive oxygen species metabolism and signaling. Ann. N. Y. Acad. Sci. 2008, 1147, 37–52. [Google Scholar] [CrossRef] [Green Version]
- Turrens, J.F. Mitochondrial formation of reactive oxygen species. J. Physiol. 2003, 552, 335–344. [Google Scholar] [CrossRef]
- Cadenas, E.; Davies, K.J. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic. Biol. Med. 2000, 29, 222–230. [Google Scholar] [CrossRef]
- Orrenius, S.; Gogvadze, V.; Zhivotovsky, B. Mitochondrial oxidative stress: Implications for cell death. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 143–183. [Google Scholar] [CrossRef] [PubMed]
- D’Autreaux, B.; Toledano, M.B. ROS as signalling molecules: Mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 2007, 8, 813–824. [Google Scholar] [CrossRef] [PubMed]
- Droge, W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002, 82, 47–95. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, A.; Gagnon, C. Formation of reactive oxygen species in spermatozoa of infertile patients. Fertil. Steril. 1992, 57, 409–416. [Google Scholar] [CrossRef]
- Zini, A.; de Lamirande, E.; Gagnon, C. Reactive oxygen species in semen of infertile patients: Levels of superoxide dismutase- and catalase-like activities in seminal plasma and spermatozoa. Int. J. Androl. 1993, 16, 183–188. [Google Scholar] [CrossRef]
- Freitas, M.J.; Vijayaraghavan, S.; Fardilha, M. Signaling mechanisms in mammalian sperm motility. Biol. Reprod. 2017, 96, 2–12. [Google Scholar] [CrossRef] [Green Version]
- St John, J.C.; Sakkas, D.; Barratt, C.L. A role for mitochondrial DNA and sperm survival. J. Androl. 2000, 21, 189–199. [Google Scholar]
- Nakada, K.; Sato, A.; Yoshida, K.; Morita, T.; Tanaka, H.; Inoue, S.; Yonekawa, H.; Hayashi, J. Mitochondria-related male infertility. Proc. Natl. Acad. Sci. USA 2006, 103, 15148–15153. [Google Scholar] [CrossRef] [Green Version]
- Ramalho-Santos, J.; Varum, S.; Amaral, S.; Mota, P.C.; Sousa, A.P.; Amaral, A. Mitochondrial functionality in reproduction: From gonads and gametes to embryos and embryonic stem cells. Hum. Reprod. Update 2009, 15, 553–572. [Google Scholar] [CrossRef] [Green Version]
- Friedman, J.R.; Nunnari, J. Mitochondrial form and function. Nature 2014, 505, 335–343. [Google Scholar] [CrossRef] [Green Version]
- Aitken, R.J.; Drevet, J.R. The Importance of Oxidative Stress in Determining the Functionality of Mammalian Spermatozoa: A Two-Edged Sword. Antioxidants 2020, 9, 111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwal, A.; Virk, G.; Ong, C.; du Plessis, S.S. Effect of oxidative stress on male reproduction. World J. Men’s Health 2014, 32, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLachlan, R.I.; de Kretser, D.M. Male infertility: The case for continued research. Med. J. Aust. 2001, 174, 116–117. [Google Scholar] [CrossRef] [PubMed]
- Pereira, R.; Sa, R.; Barros, A.; Sousa, M. Major regulatory mechanisms involved in sperm motility. Asian J. Androl. 2017, 19, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Pesini, E.; Diez, C.; Lapena, A.C.; Perez-Martos, A.; Montoya, J.; Alvarez, E.; Arenas, J.; Lopez-Perez, M.J. Correlation of sperm motility with mitochondrial enzymatic activities. Clin. Chem. 1998, 44, 1616–1620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piasecka, M.; Kawiak, J. Sperm mitochondria of patients with normal sperm motility and with asthenozoospermia: Morphological and functional study. Folia Histochem. Cytobiol. 2003, 41, 125–139. [Google Scholar] [PubMed]
- Amaral, A.; Paiva, C.; Attardo Parrinello, C.; Estanyol, J.M.; Ballesca, J.L.; Ramalho-Santos, J.; Oliva, R. Identification of proteins involved in human sperm motility using high-throughput differential proteomics. J. Proteome Res. 2014, 13, 5670–5684. [Google Scholar] [CrossRef]
- Nowicka-Bauer, K.; Lepczynski, A.; Ozgo, M.; Kamieniczna, M.; Fraczek, M.; Stanski, L.; Olszewska, M.; Malcher, A.; Skrzypczak, W.; Kurpisz, M.K. Sperm mitochondrial dysfunction and oxidative stress as possible reasons for isolated asthenozoospermia. J. Physiol. Pharmacol. 2018, 69. [Google Scholar] [CrossRef]
- St John, J.C.; Facucho-Oliveira, J.; Jiang, Y.; Kelly, R.; Salah, R. Mitochondrial DNA transmission, replication and inheritance: A journey from the gamete through the embryo and into offspring and embryonic stem cells. Hum. Reprod. Update 2010, 16, 488–509. [Google Scholar] [CrossRef] [Green Version]
- Gur, Y.; Breitbart, H. Mammalian sperm translate nuclear-encoded proteins by mitochondrial-type ribosomes. Genes Dev. 2006, 20, 411–416. [Google Scholar] [CrossRef] [Green Version]
- Bereiter-Hahn, J.; Voth, M. Dynamics of mitochondria in living cells: Shape changes, dislocations, fusion, and fission of mitochondria. Microsc. Res. Tech. 1994, 27, 198–219. [Google Scholar] [CrossRef] [PubMed]
- Vadnais, M.L.; Lin, A.M.; Gerton, G.L. Mitochondrial fusion protein MFN2 interacts with the mitostatin-related protein MNS1 required for mouse sperm flagellar structure and function. Cilia 2014, 3, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Kretser, D.M.; Loveland, K.L.; Meinhardt, A.; Simorangkir, D.; Wreford, N. Spermatogenesis. Hum. Reprod. 1998, 13 (Suppl. S1), 1–8. [Google Scholar] [CrossRef] [Green Version]
- Moreira, B.P.; Oliveira, P.F.; Alves, M.G. Molecular Mechanisms Controlled by mTOR in Male Reproductive System. Int. J. Mol. Sci. 2019, 20, 1633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hermo, L.; Pelletier, R.M.; Cyr, D.G.; Smith, C.E. Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 2: Changes in spermatid organelles associated with development of spermatozoa. Microsc. Res. Tech. 2010, 73, 279–319. [Google Scholar] [CrossRef]
- Mannella, C.A. Structure and dynamics of the mitochondrial inner membrane cristae. Biochim. Biophys. Acta 2006, 1763, 542–548. [Google Scholar] [CrossRef] [Green Version]
- De Martino, C.; Floridi, A.; Marcante, M.L.; Malorni, W.; Scorza Barcellona, P.; Bellocci, M.; Silvestrini, B. Morphological, histochemical and biochemical studies on germ cell mitochondria of normal rats. Cell Tissue Res. 1979, 196, 1–22. [Google Scholar] [CrossRef]
- Otani, H.; Tanaka, O.; Kasai, K.; Yoshioka, T. Development of mitochondrial helical sheath in the middle piece of the mouse spermatid tail: Regular dispositions and synchronized changes. Anat. Rec. 1988, 222, 26–33. [Google Scholar] [CrossRef]
- Naito, M.; Itoh, M. Patterns of infiltration of lymphocytes into the testis under normal and pathological conditions in mice. Am. J. Reprod. Immunol. 2008, 59, 55–61. [Google Scholar] [CrossRef]
- Meinhardt, A.; Hedger, M.P. Immunological, paracrine and endocrine aspects of testicular immune privilege. Mol. Cell. Endocrinol. 2011, 335, 60–68. [Google Scholar] [CrossRef]
- Boussouar, F.; Benahmed, M. Lactate and energy metabolism in male germ cells. Trends Endocrinol. Metab. 2004, 15, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Lord, T.; Nixon, B. Metabolic Changes Accompanying Spermatogonial Stem Cell Differentiation. Dev. Cell 2020, 52, 399–411. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.M.; Kwon, S.; Pak, Y.K.; Seol, H.W.; Choi, Y.M.; Park, D.J.; Park, K.S.; Lee, H.K. Dynamic changes in mitochondrial biogenesis and antioxidant enzymes during the spontaneous differentiation of human embryonic stem cells. Biochem. Biophys. Res. Commun. 2006, 348, 1472–1478. [Google Scholar] [CrossRef] [PubMed]
- Rato, L.; Alves, M.G.; Socorro, S.; Duarte, A.I.; Cavaco, J.E.; Oliveira, P.F. Metabolic regulation is important for spermatogenesis. Nat. Rev. Urol. 2012, 9, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Bajpai, M.; Gupta, G.; Setty, B.S. Changes in carbohydrate metabolism of testicular germ cells during meiosis in the rat. Eur. J. Endocrinol. 1998, 138, 322–327. [Google Scholar] [CrossRef] [Green Version]
- Courtens, J.L.; Ploen, L. Improvement of spermatogenesis in adult cryptorchid rat testis by intratesticular infusion of lactate. Biol. Reprod. 1999, 61, 154–161. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, M.; Hino, A.; Kato, J. Stimulation of protein synthesis in round spermatids from rat testes by lactate. II. Role of adenosine triphosphate (ATP). J. Biochem. 1981, 90, 933–940. [Google Scholar] [CrossRef]
- Erkkila, K.; Aito, H.; Aalto, K.; Pentikainen, V.; Dunkel, L. Lactate inhibits germ cell apoptosis in the human testis. Mol. Hum. Reprod. 2002, 8, 109–117. [Google Scholar] [CrossRef] [Green Version]
- Teslaa, T.; Teitell, M.A. Pluripotent stem cell energy metabolism: An update. EMBO J. 2015, 34, 138–153. [Google Scholar] [CrossRef]
- Xu, X.; Duan, S.; Yi, F.; Ocampo, A.; Liu, G.H.; Izpisua Belmonte, J.C. Mitochondrial regulation in pluripotent stem cells. Cell Metab. 2013, 18, 325–332. [Google Scholar] [CrossRef] [Green Version]
- Kaluz, S.; Kaluzova, M.; Stanbridge, E.J. Regulation of gene expression by hypoxia: Integration of the HIF-transduced hypoxic signal at the hypoxia-responsive element. Clin. Chim. Acta 2008, 395, 6–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.T.; Shih, Y.R.; Kuo, T.K.; Lee, O.K.; Wei, Y.H. Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem Cells 2008, 26, 960–968. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, A.D.; Beyer, M.; Krause-Buchholz, U.; Wobus, M.; Bornhauser, M.; Rodel, G. OXPHOS supercomplexes as a hallmark of the mitochondrial phenotype of adipogenic differentiated human MSCs. PLoS ONE 2012, 7, e35160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, J.; Grow, E.J.; Yi, C.; Mlcochova, H.; Maher, G.J.; Lindskog, C.; Murphy, P.J.; Wike, C.L.; Carrell, D.T.; Goriely, A.; et al. Chromatin and Single-Cell RNA-Seq Profiling Reveal Dynamic Signaling and Metabolic Transitions during Human Spermatogonial Stem Cell Development. Cell Stem Cell 2017, 21, 533–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hermann, B.P.; Cheng, K.; Singh, A.; Roa-De La Cruz, L.; Mutoji, K.N.; Chen, I.C.; Gildersleeve, H.; Lehle, J.D.; Mayo, M.; Westernstroer, B.; et al. The Mammalian Spermatogenesis Single-Cell Transcriptome, from Spermatogonial Stem Cells to Spermatids. Cell Rep. 2018, 25, 1650–1667. [Google Scholar] [CrossRef] [Green Version]
- Sohni, A.; Tan, K.; Song, H.W.; Burow, D.; de Rooij, D.G.; Laurent, L.; Hsieh, T.C.; Rabah, R.; Hammoud, S.S.; Vicini, E.; et al. The Neonatal and Adult Human Testis Defined at the Single-Cell Level. Cell Rep. 2019, 26, 1501–1517.e4. [Google Scholar] [CrossRef] [Green Version]
- Pfanner, N.; Warscheid, B.; Wiedemann, N. Mitochondrial proteins: From biogenesis to functional networks. Nat. Rev. Mol. Cell Biol. 2019, 20, 267–284. [Google Scholar] [CrossRef]
- Westermann, B. Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 2010, 11, 872–884. [Google Scholar] [CrossRef]
- Chen, H.; Vermulst, M.; Wang, Y.E.; Chomyn, A.; Prolla, T.A.; McCaffery, J.M.; Chan, D.C. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell 2010, 141, 280–289. [Google Scholar] [CrossRef] [Green Version]
- Weaver, D.; Eisner, V.; Liu, X.; Varnai, P.; Hunyady, L.; Gross, A.; Hajnoczky, G. Distribution and apoptotic function of outer membrane proteins depend on mitochondrial fusion. Mol. Cell 2014, 54, 870–878. [Google Scholar] [CrossRef] [Green Version]
- Varuzhanyan, G.; Rojansky, R.; Sweredoski, M.J.; Graham, R.L.; Hess, S.; Ladinsky, M.S.; Chan, D.C. Mitochondrial fusion is required for spermatogonial differentiation and meiosis. Elife 2019, 8. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, Q.; Wang, M.; Jiang, M.; Wang, Y.; Sun, Y.; Wang, J.; Xie, T.; Tang, C.; Tang, N.; et al. GASZ and mitofusin-mediated mitochondrial functions are crucial for spermatogenesis. EMBO Rep. 2016, 17, 220–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seitz, J.; Mobius, J.; Bergmann, M.; Meinhardt, A. Mitochondrial differentiation during meiosis of male germ cells. Int. J. Androl. 1995, 18 (Suppl. S2), 7–11. [Google Scholar]
- Reimand, J.; Isserlin, R.; Voisin, V.; Kucera, M.; Tannus-Lopes, C.; Rostamianfar, A.; Wadi, L.; Meyer, M.; Wong, J.; Xu, C.; et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 2019, 14, 482–517. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y. A statistical method for the conservative adjustment of false discovery rate (q-value). BMC Bioinform. 2017, 18, 69. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, Y.; Takahashi, K.; Okita, K.; Ichisaka, T.; Yamanaka, S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell 2009, 5, 237–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Folmes, C.D.; Nelson, T.J.; Martinez-Fernandez, A.; Arrell, D.K.; Lindor, J.Z.; Dzeja, P.P.; Ikeda, Y.; Perez-Terzic, C.; Terzic, A. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011, 14, 264–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panopoulos, A.D.; Yanes, O.; Ruiz, S.; Kida, Y.S.; Diep, D.; Tautenhahn, R.; Herrerias, A.; Batchelder, E.M.; Plongthongkum, N.; Lutz, M.; et al. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 2012, 22, 168–177. [Google Scholar] [CrossRef] [Green Version]
- Stanton, P.G. Regulation of the blood-testis barrier. Semin. Cell Dev. Biol. 2016, 59, 166–173. [Google Scholar] [CrossRef]
- Kaur, G.; Thompson, L.A.; Dufour, J.M. Sertoli cells—Immunological sentinels of spermatogenesis. Semin. Cell Dev. Biol. 2014, 30, 36–44. [Google Scholar] [CrossRef] [Green Version]
- Mital, P.; Hinton, B.T.; Dufour, J.M. The blood-testis and blood-epididymis barriers are more than just their tight junctions. Biol. Reprod. 2011, 84, 851–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, C.H.; Cheng, C.Y. The blood-testis barrier: Its biology, regulation, and physiological role in spermatogenesis. Curr. Top. Dev. Biol. 2005, 71, 263–296. [Google Scholar] [CrossRef] [PubMed]
- Jutte, N.H.; Grootegoed, J.A.; Rommerts, F.F.; van der Molen, H.J. Exogenous lactate is essential for metabolic activities in isolated rat spermatocytes and spermatids. J. Reprod. Fertil. 1981, 62, 399–405. [Google Scholar] [CrossRef]
- Riera, M.F.; Meroni, S.B.; Schteingart, H.F.; Pellizzari, E.H.; Cigorraga, S.B. Regulation of lactate production and glucose transport as well as of glucose transporter 1 and lactate dehydrogenase A mRNA levels by basic fibroblast growth factor in rat Sertoli cells. J. Endocrinol. 2002, 173, 335–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leiderman, B.; Mancini, R.E. Glycogen content in the rat testis from postnatal to adult ages. Endocrinology 1969, 85, 607–609. [Google Scholar] [CrossRef] [PubMed]
- Slaughter, G.R.; Means, A.R. Follicle-stimulating hormone activation of glycogen phosphorylase in the Sertoli cell-enriched rat testis. Endocrinology 1983, 113, 1476–1485. [Google Scholar] [CrossRef]
- Riera, M.F.; Galardo, M.N.; Pellizzari, E.H.; Meroni, S.B.; Cigorraga, S.B. Molecular mechanisms involved in Sertoli cell adaptation to glucose deprivation. Am. J. Physiol. Endocrinol. Metab. 2009, 297, E907–E914. [Google Scholar] [CrossRef] [Green Version]
- Rato, L.; Alves, M.G.; Dias, T.R.; Cavaco, J.E.; Oliveira, P.F. Testicular Metabolic Reprogramming in Neonatal Streptozotocin-Induced Type 2 Diabetic Rats Impairs Glycolytic Flux and Promotes Glycogen Synthesis. J. Diabetes Res. 2015, 2015, 973142. [Google Scholar] [CrossRef]
- Gualtieri, A.F.; Mazzone, G.L.; Rey, R.A.; Schteingart, H.F. FSH and bFGF stimulate the production of glutathione in cultured rat Sertoli cells. Int. J. Androl. 2009, 32, 218–225. [Google Scholar] [CrossRef]
- Xu, B.; Chen, M.; Ji, X.; Yao, M.; Mao, Z.; Zhou, K.; Xia, Y.; Han, X.; Tang, W. Metabolomic profiles reveal key metabolic changes in heat stress-treated mouse Sertoli cells. Toxicol. In Vitro 2015, 29, 1745–1752. [Google Scholar] [CrossRef]
- Zhang, B.B.; Zhou, G.; Li, C. AMPK: An emerging drug target for diabetes and the metabolic syndrome. Cell Metab. 2009, 9, 407–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galardo, M.N.; Riera, M.F.; Pellizzari, E.H.; Sobarzo, C.; Scarcelli, R.; Denduchis, B.; Lustig, L.; Cigorraga, S.B.; Meroni, S.B. Adenosine regulates Sertoli cell function by activating AMPK. Mol. Cell. Endocrinol. 2010, 330, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Vanderperre, B.; Cermakova, K.; Escoffier, J.; Kaba, M.; Bender, T.; Nef, S.; Martinou, J.C. MPC1-like Is a Placental Mammal-specific Mitochondrial Pyruvate Carrier Subunit Expressed in Postmeiotic Male Germ Cells. J. Biol. Chem. 2016, 291, 16448–16461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Richburg, J.H.; Younkin, S.C.; Boekelheide, K. The Fas system is a key regulator of germ cell apoptosis in the testis. Endocrinology 1997, 138, 2081–2088. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Wang, H.; Wu, H.; Chen, Y.; Han, D. Apoptotic spermatogenic cells can be energy sources for Sertoli cells. Reproduction 2009, 137, 469–479. [Google Scholar] [CrossRef] [Green Version]
- Meinhardt, A.; McFarlane, J.R.; Seitz, J.; de Kretser, D.M. Activin maintains the condensed type of mitochondria in germ cells. Mol. Cell. Endocrinol. 2000, 168, 111–117. [Google Scholar] [CrossRef]
- Li, L.; Shen, J.J.; Bournat, J.C.; Huang, L.; Chattopadhyay, A.; Li, Z.; Shaw, C.; Graham, B.H.; Brown, C.W. Activin signaling: Effects on body composition and mitochondrial energy metabolism. Endocrinology 2009, 150, 3521–3529. [Google Scholar] [CrossRef] [Green Version]
- O’Shaughnessy, P.J. Hormonal control of germ cell development and spermatogenesis. Semin. Cell Dev. Biol. 2014, 29, 55–65. [Google Scholar] [CrossRef] [PubMed]
- McLachlan, R.I.; O’Donnell, L.; Meachem, S.J.; Stanton, P.G.; de Kretser, D.M.; Pratis, K.; Robertson, D.M. Identification of specific sites of hormonal regulation in spermatogenesis in rats, monkeys, and man. Recent Prog. Horm. Res. 2002, 57, 149–179. [Google Scholar] [CrossRef]
- Fluck, C.E.; Miller, W.L.; Auchus, R.J. The 17, 20-lyase activity of cytochrome p450c17 from human fetal testis favors the delta5 steroidogenic pathway. J. Clin. Endocrinol. Metab. 2003, 88, 3762–3766. [Google Scholar] [CrossRef] [Green Version]
- Prince, F.P. Lamellar and tubular associations of the mitochondrial cristae: Unique forms of the cristae present in steroid-producing cells. Mitochondrion 2002, 1, 381–389. [Google Scholar] [CrossRef]
- Breton, S.; Ruan, Y.C.; Park, Y.J.; Kim, B. Regulation of epithelial function, differentiation, and remodeling in the epididymis. Asian J. Androl. 2016, 18, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Cornwall, G.A. New insights into epididymal biology and function. Hum. Reprod. Update 2009, 15, 213–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, Y.J.; Battistone, M.A.; Kim, B.; Breton, S. Relative contribution of clear cells and principal cells to luminal pH in the mouse epididymis. Biol. Reprod. 2017, 96, 366–375. [Google Scholar] [CrossRef]
- Shum, W.W.; Ruan, Y.C.; Da Silva, N.; Breton, S. Establishment of cell-cell cross talk in the epididymis: Control of luminal acidification. J. Androl. 2011, 32, 576–586. [Google Scholar] [CrossRef] [Green Version]
- Shum, W.W.; Da Silva, N.; McKee, M.; Smith, P.J.; Brown, D.; Breton, S. Transepithelial projections from basal cells are luminal sensors in pseudostratified epithelia. Cell 2008, 135, 1108–1117. [Google Scholar] [CrossRef] [Green Version]
- Carlin, R.W.; Lee, J.H.; Marcus, D.C.; Schultz, B.D. Adenosine stimulates anion secretion across cultured and native adult human vas deferens epithelia. Biol. Reprod. 2003, 68, 1027–1034. [Google Scholar] [CrossRef] [Green Version]
- Leung, G.P.; Wong, P.Y. Activation of cystic fibrosis transmembrane conductance regulator in rat epididymal epithelium by genistein. Biol. Reprod. 2000, 62, 143–149. [Google Scholar] [CrossRef] [Green Version]
- Brown, D.; Breton, S. Mitochondria-rich, proton-secreting epithelial cells. J. Exp. Biol. 1996, 199, 2345–2358. [Google Scholar]
- Palacios, J.; Regadera, J.; Nistal, M.; Paniagua, R. Apical mitochondria-rich cells in the human epididymis: An ultrastructural, enzymohistochemical, and immunohistochemical study. Anat. Rec. 1991, 231, 82–88. [Google Scholar] [CrossRef]
- Vernet, P.; Aitken, R.J.; Drevet, J.R. Antioxidant strategies in the epididymis. Mol. Cell. Endocrinol. 2004, 216, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Lenzi, A.; Picardo, M.; Gandini, L.; Dondero, F. Lipids of the sperm plasma membrane: From polyunsaturated fatty acids considered as markers of sperm function to possible scavenger therapy. Hum. Reprod. Update 1996, 2, 246–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aveldano, M.I.; Rotstein, N.P.; Vermouth, N.T. Lipid remodelling during epididymal maturation of rat spermatozoa. Enrichment in plasmenylcholines containing long-chain polyenoic fatty acids of the n-9 series. Biochem. J. 1992, 283 Pt 1, 235–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shalgi, R.; Seligman, J.; Kosower, N.S. Dynamics of the thiol status of rat spermatozoa during maturation: Analysis with the fluorescent labeling agent monobromobimane. Biol. Reprod. 1989, 40, 1037–1045. [Google Scholar] [CrossRef] [PubMed]
- Seligman, J.; Shalgi, R. Protein thiols in spermatozoa and epididymal fluid of rats. J. Reprod. Fertil. 1991, 93, 399–408. [Google Scholar] [CrossRef]
- Rousseaux, J.; Rousseaux-Prevost, R. Molecular localization of free thiols in human sperm chromatin. Biol. Reprod. 1995, 52, 1066–1072. [Google Scholar] [CrossRef] [Green Version]
- Chabory, E.; Damon, C.; Lenoir, A.; Henry-Berger, J.; Vernet, P.; Cadet, R.; Saez, F.; Drevet, J.R. Mammalian glutathione peroxidases control acquisition and maintenance of spermatozoa integrity. J. Anim. Sci. 2010, 88, 1321–1331. [Google Scholar] [CrossRef] [Green Version]
- Conrad, M.; Moreno, S.G.; Sinowatz, F.; Ursini, F.; Kolle, S.; Roveri, A.; Brielmeier, M.; Wurst, W.; Maiorino, M.; Bornkamm, G.W. The nuclear form of phospholipid hydroperoxide glutathione peroxidase is a protein thiol peroxidase contributing to sperm chromatin stability. Mol. Cell. Biol. 2005, 25, 7637–7644. [Google Scholar] [CrossRef] [Green Version]
- Ozkosem, B.; Feinstein, S.I.; Fisher, A.B.; O’Flaherty, C. Advancing age increases sperm chromatin damage and impairs fertility in peroxiredoxin 6 null mice. Redox Biol. 2015, 5, 15–23. [Google Scholar] [CrossRef] [Green Version]
- Ozkosem, B.; Feinstein, S.I.; Fisher, A.B.; O’Flaherty, C. Absence of Peroxiredoxin 6 Amplifies the Effect of Oxidant Stress on Mobility and SCSA/CMA3 Defined Chromatin Quality and Impairs Fertilizing Ability of Mouse Spermatozoa. Biol. Reprod. 2016, 94, 68. [Google Scholar] [CrossRef] [Green Version]
- Maser, R.L.; Magenheimer, B.S.; Calvet, J.P. Mouse plasma glutathione peroxidase. cDNA sequence analysis and renal proximal tubular expression and secretion. J. Biol. Chem. 1994, 269, 27066–27073. [Google Scholar] [CrossRef]
- Schwaab, V.; Lareyre, J.J.; Vernet, P.; Pons, E.; Faure, J.; Dufaure, J.P.; Drevet, J.R. Characterization, regulation of the expression and putative roles of two glutathione peroxidase proteins found in the mouse epididymis. J. Reprod. Fertil. Suppl. 1998, 53, 157–162. [Google Scholar] [PubMed]
- Vernet, P.; Faure, J.; Dufaure, J.P.; Drevet, J.R. Tissue and developmental distribution, dependence upon testicular factors and attachment to spermatozoa of GPX5, a murine epididymis-specific glutathione peroxidase. Mol. Reprod. Dev. 1997, 47, 87–98. [Google Scholar] [CrossRef]
- Rejraji, H.; Vernet, P.; Drevet, J.R. GPX5 is present in the mouse caput and cauda epididymidis lumen at three different locations. Mol. Reprod. Dev. 2002, 63, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Pushpa-Rekha, T.R.; Burdsall, A.L.; Oleksa, L.M.; Chisolm, G.M.; Driscoll, D.M. Rat phospholipid-hydroperoxide glutathione peroxidase. cDNA cloning and identification of multiple transcription and translation start sites. J. Biol. Chem. 1995, 270, 26993–26999. [Google Scholar] [CrossRef] [Green Version]
- Pfeifer, H.; Conrad, M.; Roethlein, D.; Kyriakopoulos, A.; Brielmeier, M.; Bornkamm, G.W.; Behne, D. Identification of a specific sperm nuclei selenoenzyme necessary for protamine thiol cross-linking during sperm maturation. FASEB J. 2001, 15, 1236–1238. [Google Scholar] [CrossRef]
- Foresta, C.; Flohe, L.; Garolla, A.; Roveri, A.; Ursini, F.; Maiorino, M. Male fertility is linked to the selenoprotein phospholipid hydroperoxide glutathione peroxidase. Biol. Reprod. 2002, 67, 967–971. [Google Scholar] [CrossRef] [Green Version]
- Fisher, A.B. Peroxiredoxin 6 in the repair of peroxidized cell membranes and cell signaling. Arch. Biochem. Biophys. 2017, 617, 68–83. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.W.; Dodia, C.; Feinstein, S.I.; Jain, M.K.; Fisher, A.B. 1-Cys peroxiredoxin, a bifunctional enzyme with glutathione peroxidase and phospholipase A2 activities. J. Biol. Chem. 2000, 275, 28421–28427. [Google Scholar] [CrossRef] [Green Version]
- Dubuisson, M.; Vander Stricht, D.; Clippe, A.; Etienne, F.; Nauser, T.; Kissner, R.; Koppenol, W.H.; Rees, J.F.; Knoops, B. Human peroxiredoxin 5 is a peroxynitrite reductase. FEBS Lett. 2004, 571, 161–165. [Google Scholar] [CrossRef] [Green Version]
- Wood, Z.A.; Poole, L.B.; Karplus, P.A. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science 2003, 300, 650–653. [Google Scholar] [CrossRef] [PubMed]
- O’Flaherty, C.; de Souza, A.R. Hydrogen peroxide modifies human sperm peroxiredoxins in a dose-dependent manner. Biol. Reprod. 2011, 84, 238–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; O’Flaherty, C. In vivo oxidative stress alters thiol redox status of peroxiredoxin 1 and 6 and impairs rat sperm quality. Asian J. Androl. 2017, 19, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Fisher, A.B.; Dodia, C.; Sorokina, E.M.; Li, H.; Zhou, S.; Raabe, T.; Feinstein, S.I. A novel lysophosphatidylcholine acyl transferase activity is expressed by peroxiredoxin 6. J. Lipid Res. 2016, 57, 587–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, P.Y.; Scarlata, E.; O’Flaherty, C. Long-Term Adverse Effects of Oxidative Stress on Rat Epididymis and Spermatozoa. Antioxidants 2020, 9, 170. [Google Scholar] [CrossRef] [Green Version]
- Chang, M.C. Fertilizing capacity of spermatozoa deposited into the fallopian tubes. Nature 1951, 168, 697–698. [Google Scholar] [CrossRef]
- Visconti, P.E.; Bailey, J.L.; Moore, G.D.; Pan, D.; Olds-Clarke, P.; Kopf, G.S. Capacitation of mouse spermatozoa. I. Correlation between the capacitation state and protein tyrosine phosphorylation. Development 1995, 121, 1129–1137. [Google Scholar]
- Visconti, P.E.; Moore, G.D.; Bailey, J.L.; Leclerc, P.; Connors, S.A.; Pan, D.; Olds-Clarke, P.; Kopf, G.S. Capacitation of mouse spermatozoa. II. Protein tyrosine phosphorylation and capacitation are regulated by a cAMP-dependent pathway. Development 1995, 121, 1139–1150. [Google Scholar]
- Galantino-Homer, H.L.; Visconti, P.E.; Kopf, G.S. Regulation of protein tyrosine phosphorylation during bovine sperm capacitation by a cyclic adenosine 3′5′-monophosphate-dependent pathway. Biol. Reprod. 1997, 56, 707–719. [Google Scholar] [CrossRef] [Green Version]
- Handrow, R.R.; First, N.L.; Parrish, J.J. Calcium requirement and increased association with bovine sperm during capacitation by heparin. J. Exp. Zool. 1989, 252, 174–182. [Google Scholar] [CrossRef]
- Ferramosca, A.; Zara, V. Bioenergetics of mammalian sperm capacitation. Biomed Res. Int 2014, 2014, 902953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrageta, D.F.; Guerra-Carvalho, B.; Sousa, M.; Barros, A.; Oliveira, P.F.; Monteiro, M.P.; Alves, M.G. Mitochondrial Activation and Reactive Oxygen-Species Overproduction during Sperm Capacitation are Independent of Glucose Stimuli. Antioxidants 2020, 9, 750. [Google Scholar] [CrossRef]
- Goodson, S.G.; Qiu, Y.; Sutton, K.A.; Xie, G.; Jia, W.; O’Brien, D.A. Metabolic substrates exhibit differential effects on functional parameters of mouse sperm capacitation. Biol. Reprod. 2012, 87, 75. [Google Scholar] [CrossRef] [PubMed]
- du Plessis, S.S.; Agarwal, A.; Mohanty, G.; van der Linde, M. Oxidative phosphorylation versus glycolysis: What fuel do spermatozoa use? Asian J. Androl. 2015, 17, 230–235. [Google Scholar] [CrossRef]
- Zhang, G.; Yang, W.; Zou, P.; Jiang, F.; Zeng, Y.; Chen, Q.; Sun, L.; Yang, H.; Zhou, N.; Wang, X.; et al. Mitochondrial functionality modifies human sperm acrosin activity, acrosome reaction capability and chromatin integrity. Hum. Reprod. 2019, 34, 3–11. [Google Scholar] [CrossRef]
- Vorup-Jensen, T.; Hjort, T.; Abraham-Peskir, J.V.; Guttmann, P.; Jensenius, J.C.; Uggerhoj, E.; Medenwaldt, R. X-ray microscopy of human spermatozoa shows change of mitochondrial morphology after capacitation. Hum. Reprod. 1999, 14, 880–884. [Google Scholar] [CrossRef] [Green Version]
- Cardullo, R.A.; Baltz, J.M. Metabolic regulation in mammalian sperm: Mitochondrial volume determines sperm length and flagellar beat frequency. Cell Motil. Cytoskelet. 1991, 19, 180–188. [Google Scholar] [CrossRef]
- Leclerc, P.; de Lamirande, E.; Gagnon, C. Cyclic adenosine 3′,5′monophosphate-dependent regulation of protein tyrosine phosphorylation in relation to human sperm capacitation and motility. Biol. Reprod. 1996, 55, 684–692. [Google Scholar] [CrossRef]
- Dimitriadis, F.; Giannakis, D.; Pardalidis, N.; Zikopoulos, K.; Paraskevaidis, E.; Giotitsas, N.; Kalaboki, V.; Tsounapi, P.; Baltogiannis, D.; Georgiou, I.; et al. Effects of phosphodiesterase-5 inhibitors on sperm parameters and fertilizing capacity. Asian J. Androl. 2008, 10, 115–133. [Google Scholar] [CrossRef]
- Griveau, J.F.; Renard, P.; Le Lannou, D. Superoxide anion production by human spermatozoa as a part of the ionophore-induced acrosome reaction process. Int. J. Androl. 1995, 18, 67–74. [Google Scholar] [CrossRef]
- Goldman, R.; Ferber, E.; Zort, U. Reactive oxygen species are involved in the activation of cellular phospholipase A2. FEBS Lett. 1992, 309, 190–192. [Google Scholar] [CrossRef] [Green Version]
- Du Plessis, S.S.; Agarwal, A.; Halabi, J.; Tvrda, E. Contemporary evidence on the physiological role of reactive oxygen species in human sperm function. J. Assist. Reprod. Genet. 2015, 32, 509–520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Lamirande, E.; Gagnon, C. Impact of reactive oxygen species on spermatozoa: A balancing act between beneficial and detrimental effects. Hum. Reprod. 1995, 10 (Suppl. S1), 15–21. [Google Scholar] [CrossRef] [PubMed]
- Ford, W.C. Regulation of sperm function by reactive oxygen species. Hum. Reprod. Update 2004, 10, 387–399. [Google Scholar] [CrossRef] [PubMed]
- de Lamirande, E.; Gagnon, C. Reactive oxygen species and human spermatozoa. I. Effects on the motility of intact spermatozoa and on sperm axonemes. J. Androl. 1992, 13, 368–378. [Google Scholar] [PubMed]
- de Lamirande, E.; Gagnon, C. Reactive oxygen species and human spermatozoa. II. Depletion of adenosine triphosphate plays an important role in the inhibition of sperm motility. J. Androl. 1992, 13, 379–386. [Google Scholar]
- De Kretser, D.M.; Baker, H.W. Infertility in men: Recent advances and continuing controversies. J. Clin. Endocrinol. Metab. 1999, 84, 3443–3450. [Google Scholar] [CrossRef]
- Xuan, W.; Lamhonwah, A.M.; Librach, C.; Jarvi, K.; Tein, I. Characterization of organic cation/carnitine transporter family in human sperm. Biochem. Biophys. Res. Commun. 2003, 306, 121–128. [Google Scholar] [CrossRef]
- Liu, F.J.; Liu, X.; Han, J.L.; Wang, Y.W.; Jin, S.H.; Liu, X.X.; Liu, J.; Wang, W.T.; Wang, W.J. Aged men share the sperm protein PATE1 defect with young asthenozoospermia patients. Hum. Reprod. 2015, 30, 861–869. [Google Scholar] [CrossRef] [Green Version]
- Shivaji, S.; Kota, V.; Siva, A.B. The role of mitochondrial proteins in sperm capacitation. J. Reprod. Immunol. 2009, 83, 14–18. [Google Scholar] [CrossRef]
- Asquith, K.L.; Harman, A.J.; McLaughlin, E.A.; Nixon, B.; Aitken, R.J. Localization and significance of molecular chaperones, heat shock protein 1, and tumor rejection antigen gp96 in the male reproductive tract and during capacitation and acrosome reaction. Biol. Reprod. 2005, 72, 328–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, Y.J.; Kwon, W.S.; Oh, S.A.; Pang, M.G. Fertility-Related Proteomic Profiling Bull Spermatozoa Separated by Percoll. J. Proteome Res. 2012, 11, 4162–4168. [Google Scholar] [CrossRef] [PubMed]
- Kwon, W.S.; Oh, S.A.; Kim, Y.J.; Rahman, M.S.; Park, Y.J.; Pang, M.G. Proteomic approaches for profiling negative fertility markers in inferior boar spermatozoa. Sci. Rep. 2015, 5, 13821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bansal, S.K.; Gupta, N.; Sankhwar, S.N.; Rajender, S. Differential Genes Expression between Fertile and Infertile Spermatozoa Revealed by Transcriptome Analysis. PLoS ONE 2015, 10, e0127007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feugang, J.M.; Rodriguez-Osorio, N.; Kaya, A.; Wang, H.; Page, G.; Ostermeier, G.C.; Topper, E.K.; Memili, E. Transcriptome analysis of bull spermatozoa: Implications for male fertility. Reprod. Biomed. Online 2010, 21, 312–324. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Heredia, J.; de Mateo, S.; Vidal-Taboada, J.M.; Ballesca, J.L.; Oliva, R. Identification of proteomic differences in asthenozoospermic sperm samples. Hum. Reprod. 2008, 23, 783–791. [Google Scholar] [CrossRef] [Green Version]
- Parte, P.P.; Rao, P.; Redij, S.; Lobo, V.; D’Souza, S.J.; Gajbhiye, R.; Kulkarni, V. Sperm phosphoproteome profiling by ultra performance liquid chromatography followed by data independent analysis (LC-MS(E)) reveals altered proteomic signatures in asthenozoospermia. J. Proteom. 2012, 75, 5861–5871. [Google Scholar] [CrossRef]
- Kwon, W.S.; Rahman, M.S.; Ryu, D.Y.; Park, Y.J.; Pang, M.G. Increased male fertility using fertility-related biomarkers. Sci. Rep. 2015, 5, 15654. [Google Scholar] [CrossRef] [Green Version]
- Kwon, W.S.; Rahman, M.S.; Lee, J.S.; Yoon, S.J.; Park, Y.J.; Pang, M.G. Discovery of predictive biomarkers for litter size in boar spermatozoa. Mol. Cell. Proteom. 2015, 14, 1230–1240. [Google Scholar] [CrossRef] [Green Version]
- Moscatelli, N.; Lunetti, P.; Braccia, C.; Armirotti, A.; Pisanello, F.; De Vittorio, M.; Zara, V.; Ferramosca, A. Comparative Proteomic Analysis of Proteins Involved in Bioenergetics Pathways Associated with Human Sperm Motility. Int. J. Mol. Sci. 2019, 20, 3000. [Google Scholar] [CrossRef] [Green Version]
- Panner Selvam, M.K.; Agarwal, A.; Pushparaj, P.N.; Baskaran, S.; Bendou, H. Sperm Proteome Analysis and Identification of Fertility-Associated Biomarkers in Unexplained Male Infertility. Genes 2019, 10, 522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwal, A.; Sharma, R.; Samanta, L.; Durairajanayagam, D.; Sabanegh, E. Proteomic signatures of infertile men with clinical varicocele and their validation studies reveal mitochondrial dysfunction leading to infertility. Asian J. Androl. 2016, 18, 282–291. [Google Scholar] [CrossRef] [PubMed]
- Hosseinifar, H.; Gourabi, H.; Salekdeh, G.H.; Alikhani, M.; Mirshahvaladi, S.; Sabbaghian, M.; Modarresi, T.; Gilani, M.A. Study of sperm protein profile in men with and without varicocele using two-dimensional gel electrophoresis. Urology 2013, 81, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Agarwal, A.; Mohanty, G.; Hamada, A.J.; Gopalan, B.; Willard, B.; Yadav, S.; du Plessis, S. Proteomic analysis of human spermatozoa proteins with oxidative stress. Reprod. Biol. Endocrinol. 2013, 11, 48. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Wu, Y.; Jin, K.; Lu, H.; Liu, F.; Guo, Y.; Yan, F.; Shi, W.; Liu, Y.; Cao, X.; et al. Differential proteomic profiling in human spermatozoa that did or did not result in pregnancy via IVF and AID. Proteom. Clin. Appl. 2013, 7, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Stoeckli, E.T. Understanding axon guidance: Are we nearly there yet? Development 2018, 145. [Google Scholar] [CrossRef] [Green Version]
- Clavier, A.; Rincheval-Arnold, A.; Colin, J.; Mignotte, B.; Guenal, I. Apoptosis in Drosophila: Which role for mitochondria? Apoptosis 2016, 21, 239–251. [Google Scholar] [CrossRef]
- Nakajima, Y.I.; Kuranaga, E. Caspase-dependent non-apoptotic processes in development. Cell Death Differ. 2017, 24, 1422–1430. [Google Scholar] [CrossRef]
- Moore, S.W.; Kennedy, T.E. Protein kinase A regulates the sensitivity of spinal commissural axon turning to netrin-1 but does not switch between chemoattraction and chemorepulsion. J. Neurosci. 2006, 26, 2419–2423. [Google Scholar] [CrossRef] [Green Version]
- Moore, S.W.; Lai Wing Sun, K.; Xie, F.; Barker, P.A.; Conti, M.; Kennedy, T.E. Soluble adenylyl cyclase is not required for axon guidance to netrin-1. J. Neurosci. 2008, 28, 3920–3924. [Google Scholar] [CrossRef]
- Ford, W.C. Glycolysis and sperm motility: Does a spoonful of sugar help the flagellum go round? Hum. Reprod. Update 2006, 12, 269–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, R.M. Moving to the beat: A review of mammalian sperm motility regulation. Reprod. Fertil. Dev. 2006, 18, 25–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giles, R.E.; Blanc, H.; Cann, H.M.; Wallace, D.C. Maternal inheritance of human mitochondrial DNA. Proc. Natl. Acad. Sci. USA 1980, 77, 6715–6719. [Google Scholar] [CrossRef] [Green Version]
- Cann, R.L.; Stoneking, M.; Wilson, A.C. Mitochondrial DNA and human evolution. Nature 1987, 325, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, M.; Vissing, J. Paternal inheritance of mitochondrial DNA. N. Engl. J. Med. 2002, 347, 576–580. [Google Scholar] [CrossRef] [PubMed]
- Shitara, H.; Hayashi, J.I.; Takahama, S.; Kaneda, H.; Yonekawa, H. Maternal inheritance of mouse mtDNA in interspecific hybrids: Segregation of the leaked paternal mtDNA followed by the prevention of subsequent paternal leakage. Genetics 1998, 148, 851–857. [Google Scholar] [PubMed]
- Cummins, J.M. Fertilization and elimination of the paternal mitochondrial genome. Hum. Reprod. 2000, 15 (Suppl. S2), 92–101. [Google Scholar] [CrossRef] [Green Version]
- DeLuca, S.Z.; O’Farrell, P.H. Barriers to male transmission of mitochondrial DNA in sperm development. Dev. Cell 2012, 22, 660–668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, S.M.; Ge, Z.J.; Wang, Z.W.; Jiang, Z.Z.; Wang, Z.B.; Ouyang, Y.C.; Hou, Y.; Schatten, H.; Sun, Q.Y. Unique insights into maternal mitochondrial inheritance in mice. Proc. Natl. Acad. Sci. USA 2013, 110, 13038–13043. [Google Scholar] [CrossRef] [Green Version]
- Sutovsky, P. Ubiquitin-dependent proteolysis in mammalian spermatogenesis, fertilization, and sperm quality control: Killing three birds with one stone. Microsc. Res. Tech. 2003, 61, 88–102. [Google Scholar] [CrossRef]
- Song, W.H.; Yi, Y.J.; Sutovsky, M.; Meyers, S.; Sutovsky, P. Autophagy and ubiquitin-proteasome system contribute to sperm mitophagy after mammalian fertilization. Proc. Natl. Acad. Sci. USA 2016, 113, E5261–E5270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sutovsky, P.; Moreno, R.D.; Ramalho-Santos, J.; Dominko, T.; Simerly, C.; Schatten, G. Ubiquitin tag for sperm mitochondria. Nature 1999, 402, 371–372. [Google Scholar] [CrossRef] [PubMed]
- Thompson, W.E.; Ramalho-Santos, J.; Sutovsky, P. Ubiquitination of prohibitin in mammalian sperm mitochondria: Possible roles in the regulation of mitochondrial inheritance and sperm quality control. Biol. Reprod. 2003, 69, 254–260. [Google Scholar] [CrossRef] [PubMed]




| Highly Expressed Genes in Spermatogonia | Highly Expressed Genes in Spermatocytes and Spermatids |
|---|---|
| 22-oxoglutarate dehydrogenase-like, mitochondrial (OGDHL); 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 (PFKFB3); Aminoacyl tRNA synthase complex-interacting multifunctional protein 1 (AIMP1), ATP-dependent 6-phosphofructokinase, liver type (PFKL); ATP-dependent 6-phosphofructokinase, muscle type (PFKM); Enolase (ENO) 3; ENO4, Fructose-bisphosphate aldolase A (ALDOA); GDH/6PGL endoplasmic bifunctional protein (H6PD); Glyceraldehyde-3-phosphate dehydrogenase (GAPDH); Glycosylphosphatidylinositol (GPI); Myc proto-oncogene protein (MYC); Nischarin (NISCH); Phosphoglucomutase-2 (PGM2); Phosphoglycerate mutase 1 (PGAM1); Pyruvate dehydrogenase (acetyl-transferring) kinase isozyme 2, mitochondrial (PDK2); Pyruvate dehydrogenase E1 component subunit beta, mitochondrial (PDHB); Triosephosphate isomerase (TPI1) | 39S ribosomal protein L53, mitochondrial (MRPL53); Complex I assembly factor ACAD9, mitochondrial (ACAD9); Cytosolic carboxypeptidase 1 (AGTPBP1); Apoptosis-inducing factor 1, mitochondrial (AIFM1); Polyamine-transporting ATPase 13A2 (ATP13A2); ATP synthase subunit epsilon, mitochondrial (ATP5E); ATP5G2; ATP5J; ATP5L; Aurora kinase A-interacting protein (AURKAIP1); Mitochondrial chaperone BCS1 (BCS1L); Coiled-coil-helix-coiled-coil-helix domain-containing protein 5 (CHCHD5); Clustered mitochondria protein homolog (CLUH); Cytochrome c oxidase assembly factor 1 homolog (COA1); COA5; Cytochrome c oxidase copper chaperone COX19 (COX19); COX20; COX5A; COX6C; COX7B; COX17; Cytochrome c oxidase subunit NDUFA4 (NDUFA4); 28S ribosomal protein S29, mitochondrial (DAP3); DnaJ homolog subfamily A member 3, mitochondrial (DNAJA3); Mitochondrial fission 1 protein (FIS1); Frataxin, mitochondrial (FXN); Elongation factor G, mitochondrial (GFM1); GFM2; Golgi phosphoprotein 3 (GOLPH3); Histidine—tRNA ligase, cytoplasmic 1 (HARS1); HARS2; Probable leucine—tRNA ligase, mitochondrial (LARS2); 39S ribosomal protein L1, mitochondrial (MRPL1); MRPL14; MRPL18; MRPL19; MRPL20; MRPL24; MRPL3; MRPL30; MRPL44; MRPL47; MRPL55; 28S ribosomal protein S18b, mitochondrial (MRPS10); MRPS14; MRPS15; MRPS17; MRPS18A; MRPS18C; MRPS21; MRPS33; MRPS5; Transcription termination factor 4, mitochondrial (MTERF4); Mitochondrial ubiquitin ligase activator of NFKB 1 (MUL1); NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 1 (NDUFA1); NDUFA11; NDUFA12; NDUFA13; NDUFA2; NDUFA3; NDUFA4; NDUFA5; NDUFA6; NDUFA7; NDUFAF2; NDUFAF3; NDUFAF7; NDUFB1; NDUFB2; NDUFB3; NDUFB4; NDUFB7; NDUFB8; NDUFC1; NDUFC2; NDUFS1; NDUFS2; NDUFS5; NDUFS6; NDUFS7; Nitric oxide-associated protein 1 (NOA1); Presenilins-associated rhomboid-like protein, mitochondrial (PARL); Glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial (QRSL1); Small integral membrane protein 20 (SMIM20); Sequestosome-1 (SQSTM1); Dimethyladenosine transferase 2, mitochondrial (TFB2M); Ubiquinol-cytochrome-c reductase complex assembly factor 2 (UQCC2); Cytochrome b-c1 complex subunit 9 (UQCR10) |
| Molecular Functions | FDR | Related Genes | |
|---|---|---|---|
| Highly expressed genes in Spermatogonia | Glycolysis | <0.001 | GPI, TPI1, PGAM1, PFKL, PFKFB3, ENO3, PFKM, GAPDH |
| Glucose metabolism | <0.001 | GPI, TPI1, PGAM1, PFKL, PFKFB3, ENO3, PFKM, GAPDH | |
| Metabolism of carbohydrates | <0.001 | GPI, TPI1, PGAM1, PFKL, PFKFB3, ENO3, PFKM, GAPDH, PGM2 | |
| Gluconeogenesis | <0.001 | GPI, TPI1, PGAM1, ENO3, GAPDH | |
| Metabolism | <0.001 | GPI, TPI1, PGAM1, PFKL, PFKFB3, ENO3, PFKM, GAPDH, PGM2, PDK2, AIMP1, PDHB | |
| Regulation of pyruvate dehydrogenase (PDH) complex | <0.05 | PDK2, PDHB | |
| Highly expressed genes in Spermatocytes and Spermatids | Respiratory electron transport | <0.001 | NDUFA4, NDUFS5, NDUFB7, NDUFA6, NDUFC2, NDUFA13, NDUFC1, NDUFA12, NDUFB1, NDUFB2, NDUFB3, NDUFA5, NDUFB4, NDUFA2, NDUFAF7, NDUFA1, NDUFAF3, NDUFS6, ACAD9, NDUFS2, NDUFS1, NDUFS7, NDUFA3, NDUFA7, NDUFAF2, NDUFA11, UQCR10, COX20, COX7B, COX5A, COX6C |
| Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins | <0.001 | NDUFA4, NDUFS5, NDUFB7, NDUFA6, NDUFC2, NDUFA13, NDUFC1, NDUFA12, NDUFB1, NDUFB2, NDUFB3, NDUFA5, NDUFB4, NDUFA2, NDUFAF7, NDUFA1, NDUFAF3, NDUFS6, ACAD9, NDUFS2, NDUFS1, NDUFS7, NDUFA3, NDUFA7, NDUFAF2, NDUFA11, UQCR10, COX20, COX7B, COX5A, COX6C, ATP5J | |
| Complex I biogenesis | <0.001 | NDUFS5, NDUFB7, NDUFA6, NDUFC2, NDUFA13, NDUFC1, NDUFA12, NDUFB1, NDUFB2, NDUFB3, NDUFA5, NDUFB4, NDUFA2, NDUFAF7, NDUFA1, NDUFAF3, NDUFS6, ACAD9, NDUFS2, NDUFS1, NDUFS7, NDUFA3, NDUFA7, NDUFAF2, NDUFA11 | |
| The citric acid (TCA) cycle and respiratory electron transport | <0.001 | NDUFA4, NDUFS5, NDUFB7, NDUFA6, NDUFC2, NDUFA13, NDUFC1, NDUFA12, NDUFB1, NDUFB2, NDUFB3, NDUFA5, NDUFB4, NDUFA2, NDUFAF7, NDUFA1, NDUFAF3, NDUFS6, ACAD9, NDUFS2, NDUFS1, NDUFS7, NDUFA3, NDUFA7, NDUFAF2, NDUFA11, UQCR10, COX20, COX7B, COX5A, COX6C, ATP5J | |
| Mitochondrial translation | <0.001 | MRPL53, AURKAIP1, GFM2, DAP3, MRPS17, MRPS18C, MRPS15, MRPS18A, MRPS21, MRPL47, MRPL1, MRPL3, MRPS33, MRPS14, MRPS10, MRPS5, MRPL30, MRPL20, MRPL24, MRPL14, GFM1, MRPL19, MRPL18, MRPL55, MRPL44 | |
| Mitochondrial translation termination | <0.001 | MRPL53, AURKAIP1, GFM2, DAP3, MRPS17, MRPS18C, MRPS15, MRPS18A, MRPS21, MRPL47, MRPL1, MRPL3, MRPS33, MRPS14, MRPS10, MRPS5, MRPL30, MRPL20, MRPL24, MRPL14, MRPL19, MRPL18, MRPL55, MRPL44 | |
| Mitochondrial translation elongation | <0.001 | MRPL53, AURKAIP1, DAP3, MRPS17, MRPS18C, MRPS15, MRPS18A, MRPS21, MRPL47, MRPL1, MRPL3, MRPS33, MRPS14, MRPS10, MRPS5, MRPL30, MRPL20, MRPL24, MRPL14, GFM1, MRPL19, MRPL18, MRPL55, MRPL44 | |
| Mitochondrial translation initiation | <0.001 | MRPL53, AURKAIP1, DAP3, MRPS17, MRPS18C, MRPS15, MRPS18A, MRPS21, MRPL47, MRPL1, MRPL3, MRPS33, MRPS14, MRPS10, MRPS5, MRPL30, MRPL20, MRPL24, MRPL14, MRPL19, MRPL18, MRPL55, MRPL44 | |
| Translation | <0.001 | MRPL53, AURKAIP1, GFM2, DAP3, MRPS17, MRPS18C, MRPS15, MRPS18A, HARS2, MRPS21, MRPL47, LARS2, MRPL1, MRPL3, MRPS33, MRPS14, MRPS10, MRPS5, MRPL30, MRPL20, MRPL24, MRPL14, GFM1, MRPL19, MRPL18, MRPL55, MRPL44 | |
| Metabolism | <0.001 | NDUFA4, NDUFS5, NDUFB7, NDUFA6, NDUFC2, NDUFA13, NDUFC1, NDUFA12, NDUFB1, NDUFB2, NDUFB3, NDUFA5, NDUFB4, NDUFA2, NDUFAF7, NDUFA1, NDUFAF3, NDUFS6, ACAD9, NDUFS2, NDUFS1, NDUFS7, NDUFA3, NDUFA7, NDUFAF2, NDUFA11, UQCR10, COX20, COX7B, COX5A, COX6C, ATP5J | |
| Metabolism of proteins | <0.001 | MRPL53, AURKAIP1, GFM2, DAP3, MUL1, AGTPBP1, MRPS17, MRPS18C, MRPS15, MRPS18A, HARS2, MRPS21, MRPL47, LARS2, MRPL1, MRPL3, MRPS33, MRPS14, MRPS10, MRPS5, MRPL30, MRPL20, MRPL24, MRPL14, GFM1, MRPL19, MRPL18, MRPL55, MRPL44 |
| Highly Expressed Proteins in Spermatozoa from Fertile Men | Highly Expressed Proteins in Spermatozoa from Fertile Men | Pathological Condition |
|---|---|---|
| CSE1 chromosome segregation 1-like protein (CSE1L); Beta actin (ACTB); Chaperonin containing TCP1, subunit 8 (theta); Clathrin heavy chain 1 (CLTC); Enolase 1 (ENO1); Eukaryotic translation elongation factor 2 (EEF2); Fibronectin 1 isoform 3 preproprotein (FN1); Heat shock 70 kda protein 2 (HSPA2); Heat shock 90 kda protein 1, alpha isoform 1 (HSP90AA1); Heat shock protein beta-1 (HSPB1); L-lactate dehydrogenase C (LDHC); Peptidylprolyl isomerase A (PPIA); Peroxiredoxin 6 (PRDX6); Phosphoglycerate kinase 2 (PGK2); Prolactin-induced protein (PIP); Prostate specific antigen isoform 1 preproprotein (KLK3); Pyruvate kinase, muscle isoform M2 (PKM); Semenogelin I isoform a preproprotein (SEMG1); Semenogelin II precursor (SEMG2); Sorbitol dehydrogenase (SORD); Tubulin, beta 4 (TUBB4A); Tyrosine 3/tryptophan 5 -monooxygenase activation protein, zeta polypeptide (YWHAZ); Valosin-containing protein (VCP) | Acetyl-Coenzyme A acetyltransferase 1 precursor (ACAT1); Acid phosphatase, prostate short isoform precursor (ACPP); Angiotensin I converting enzyme 1 isoform 1 precursor (ACE); ATP synthase, H + transporting, mitochondrial F1 complex, alpha subunit precursor (ATP5A1); Brain creatine kinase (CKB); Chaperonin containing TCP1, subunit 4 (CCT4); Chaperonin containing TCP1, subunit 5 (CCT8); Chromosome 20 open reading frame 3 (c20orf3); Clusterin preproprotein (CLU); Eukaryotic translation elongation factor 1 alpha 1 (EEF1A1); Fru fumarate hydratase precursor (FH); Glutamine synthetase (GLUL); Glutathione peroxidase 4 isoform A precursor (GPX4); Glutathione S-transferase mu 3 (GSTM3); Glyceraldehyde-3-phosphate dehydrogenase (GAPDH); Heat shock 70 kda protein 5 (HSPA5); Heat shock protein 90 kda beta, member 1 (HSP90B1); Histone cluster 1, h2aa (HIST1H2AA); Histone cluster 1, h2ba (HIST1H2BA); Lactotransferrin precursor (LTF); Mitochondrial ATP synthase beta subunit precursor (ATP5B); Mitochondrial malate dehydrogenase precursor (MDH2); Olfactomedin 4 precursor (OLFM4); Outer dense fiber of sperm tails 2 isoform 2 (ODF2); Phosphoglycerate dehydrogenase (PHGDH); Phospholipase A2, group IIA precursor (PLA2G2A); Protein disulfide-isomerase A3 precursor (PDIA3); RAB2A, member RAS oncogene family (RAB2A); Ropporin (ROPN1); Ropporin, rhophilin associated protein 1B (ROPN1B); Saccharopine dehydrogenase (SCCPDH); Sperm autoantigenic protein 17 (SPA17); Sperm protein associated with the nucleus, X chromosome, family member C (SPANX); Transglutaminase 4 (TGM4); Triosephosphate isomerase 1 isoform 1 (TPI1); Tubulin alpha 6 (TUBA1C); Tubulin, alpha 3c (TUBA3C); Tubulin, beta, 2 (TUBB4B); Voltage-dependent anion channel 2 (VDAC2) | Oxidative stress [164] |
| AIG2-like domain 1(A2LD1); EF-hand calcium binding domain (RCN2); F-box protein 2 (FBXO2); Cell division cycle 34 homolog, ubiquitin-conjugating enzyme E2R2 (CDC34, UBE2R2); Calumenin (CALU); Regulator of chromosome condensation 1(RCC1); ATPase, Na+/K+ transporting, beta 3 polypeptide (ATP1B3); Progesterone receptor membrane component 1/2 (PGRMC1/ PGRMC2); Polyribonucleotide nucleotidyltransferase 1(PNPT1); Acyl-coa thioesterase 7(ACOT7); Acrosomal vesicle protein 1(ACRV1); Fragile X mental retardation 1 neighbor (FMR1NB); Cysteine-rich secretory protein 2/3 (CRISP2/CRISP3) | Actin, alpha 1, skeletal muscle (ACTA1); Abhydrolase domain containing 2 (ABHD2); Phosphofructokinase, muscle (PFKM); Beta-2-microglobulin (B2M); Protein phosphatase 5(PPP5C) | IVF failed [165] |
| Clusterin (CLU); RNA-binding regulatory subunit of oncogene DJ1 (PARK7); Prostate-specific antigen (KLK3); Prolactin induced protein (PIP); Superoxide dismutase 1 (SOD1); Semenogelin-2 (SEMG2); ATP synthase subunit δ, mitochondrial (ATP5D); 78-kda glucose-regulated protein precursor (HSPA5); Aspartate-rich protein 1 (DRICH1); Nucleoporin p58/p45 (NUP58); Protein c9orf135 (c9orf135); Coiled-coil domain-containing protein 42 (CCDC42); 5’-deoxynucleotidase HDDC2 (HDDC2); Protein DPCD (DPCD); Heterogeneous nuclear ribonucleoprotein M (HNRNPM); Syntaxin-12 (STX12); Acrosin-binding protein (ACRBP); Adenylate kinase 7 (AK7); Sodium/potassium-transporting atpase subunit alpha-4 (ATP1A4); T-complex protein 1 subunit zeta-2 (CCT6B); Heat shock 70kda protein 2, isoform CRA_a (HSPA2); Heat shock 70 kda protein 4L (HSPA4L); Nucleoside diphosphate kinase (NME5); Ruvb-like helicase (EC 3.6.4.12); (RUVBL1); Sperm autoantigenic protein 17 (SPA17); Camp-dependent protein kinase catalytic subunit alpha (PRKACA); Truncated camp-dependent protein kinase A type 1A regulatory subunit (PRKAR1A); Aconitate hydratase, mitochondrial (ACO2); NADH-ubiquinone oxidoreductase 75 kda subunit, mitochondrial (NDUFS1); 2-oxoglutarate dehydrogenase, mitochondrial (OGDH); Cytochrome b-c1 complex subunit 2, mitochondrial (UQCRC2); Apolipoprotein A-I, isoform CRA_a (APOA1); V-type proton atpase subunit E 1 (ATP6V1E1); Isocitrate dehydrogenase [NAD] subunit, mitochondrial (IDH3B I); 2-oxoglutarate dehydrogenase, mitochondrial (ODO1); Short-chain acyl-coa dehydrogenase (ACADS); Long-chain-fatty-acid--coa ligase 6 (ACSL6); Delta(3,5); -Delta(2,4); -dienoyl-coa isomerase, mitochondrial (ECH1); Voltage-dependent calcium channel subunit alpha-2/delta-2 (CACNA2D2); Glucosidase, alpha acid (Pompe disease, glycogen storage disease type II); isoform CRA_a (GAA); cAMP-dependent protein kinase type I-alpha regulatory subunit (PRKAR1A) | Gelsolin (GSN); Prostatic acid phosphatase precursor (ACPP); Integrin alpha-M (ITAM); Integrin beta-2 (FINC); Outer dense fiber protein 2 (ODFP2); Protein-glutamine gamma-glutamyltransferase 4 (TGM4); Filamin-B (FLNB); Tektin-3 (TEKT3); Cytosolic non-specific dipeptidase (CNDP2); Ras gtpase-activating-like protein IQGAP1 (p195); (IQGA1); Azurocidin (CAP7); ATP- citrate synthase (ACLY); Glutamine--fructose-6-phosphate aminotransferase (GFPT1) | Varicocele [162,163] |
| Cullin-associated NEDD8-dissociated protein 1 (CAND1); Endoplasmin precursor (ENPL); Sperm equatorial segment protein 1 precursor (SPESP); Semenogelin-2 precursor (SEMG2); Semenogelin-1 preproprotein (SEMG1); 3’(2’); 5’-bisphosphate nucleotidase 1 isoform X3 (BPNT1); Mitochondrial import receptor subunit TOM40 homolog (TOM40); Protein FAM71B (FA71B); L-amino-acid oxidase isoform 2 precursor (OXLA); Izumo sperm-egg fusion protein 3 precursor (S4R3E6); Sperm surface protein Sp17 (SP17); Diablo homolog, mitochondrial isoform 1 precursor (DBLOH); Solute carrier family 2, facilitated glucose transporter member 14 isoform a (GTR14); Mitochondrial pyruvate carrier 2 isoform X1 (MPC2); Multifunctional protein ADE2 isoform 2 (PUR6); Thioredoxin domain-containing protein 3 (TXND3); Plasma serine protease inhibitor preproprotein (IPSP); Mannose-6-phosphate isomerase isoform 1 (MPI); NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 11 isoform 1 (NDUAB); UV excision repair protein RAD23 homolog B isoform 1 (RD23B); Cysteine-rich secretory protein 1 isoform 1 precursor (CRIS1); Four and a half LIM domains protein 1 isoform 5 (FHL1); ADP-ribosylation factor 1 (ARF1); Fibronectin isoform 1 preproprotein (Q6MZM7); 3-mercaptopyruvate sulfurtransferase isoform 1 (THTM); Leucine zipper transcription factor-like protein 1 isoform 1 (LZTL1); Glucosamine-6-phosphate isomerase 2 isoform 1 (GNPI2); Calcium-binding mitochondrial carrier protein Aralar2 isoform 1 (CMC2); Dynactin subunit 2 isoform 2 (DCTN2); Mimitin, mitochondrial (NDUF2); 26S protease regulatory subunit 7 isoform 1 (PRS7); Ubiquitin carboxyl-terminal hydrolase isozyme L1 (UCHL1); Protein phosphatase 1A isoform 3 (PPM1A); 26S protease regulatory subunit 6A (PRS6A); Inactive serine protease 54 precursor (PRS54); Valine--trna ligase (SYVC); EGF-like repeat and discoidin I-like domain-containing protein 3 isoform 1 precursor (EDIL3); Acyl-coa dehydrogenase family member 9, mitochondrial (ACAD9); Matrix-remodeling-associated protein 5 precursor (MXRA5); Lon protease homolog, mitochondrial isoform 1 precursor (LONM); Redox-regulatory protein FAM213A isoform 1 precursor (PXL2A); Translocation protein SEC63 homolog (SEC63); Alpha-soluble NSF attachment protein (SNAA); Exportin-2 isoform 1 (XPO2); Dehydrogenase/reductase SDR family member 7B (DRS7B); Calcium-binding mitochondrial carrier protein Aralar1 (CMC1); SUN domain-containing protein 3 isoform 2 (SUN3); 26S protease regulatory subunit 8 isoform 1 (PRS8); Tetratricopeptide repeat protein 25 (TTC25); Casein kinase I isoform alpha isoform 3 (KC1A); Dynactin subunit 1 isoform 5 (DCTN1); NADPH--cytochrome P450 reductase (NCPR); Syntaxin-12 (STX12); Nucleoporin NUP53 isoform a (NUP35); Extracellular matrix protein 1 isoform 1 precursor (ECM1); Ribonuclease 4 precursor (RNAS4); Calcium/calmodulin-dependent protein kinase type IV (KCC4); Fibronectin isoform 3 preproprotein (Q6MZF4); Maestro heat-like repeat-containing protein family member 7 (MROH7); Short/branched chain specific acyl-coa dehydrogenase, mitochondrial precursor (ACDSB); Choline transporter-like protein 5 isoform B (CTL5); 26S protease regulatory subunit 6B isoform 1 (PRS6B); Protein canopy homolog 3 precursor (CNPY3); Carboxypeptidase B preproprotein (CBPB1); CMT1A duplicated region transcript 15 protein (CDRTF); Dolichyldiphosphatase 1 isoform a (DOPP1); Serotransferrin precursor (TRFE); Acetyl-coa acetyltransferase, cytosolic (THIC); Mitochondrial coenzyme A transporter SLC25A42 isoform X1 (S2542); Arrestin domain-containing protein 5 (ARRD5); Aquaporin-7 (AQP7) | Protein NDRG1 (NDRG1); Laminin subunit gamma-1 precursor (LAMC1); Succinyl-coa:3-ketoacid coenzyme A transferase 2, mitochondrial precursor (SCOT2); Cytosolic non-specific dipeptidase isoform X2 (CNDP2); Testis-specific serine kinase substrate (TSKS); Matrix metalloproteinase-9 preproprotein (MMP9); Myoferlin isoform b (MYOF); Alpha-actinin-1 isoform c (ACTN1); Prostate stem cell antigen preproprotein (D3DWI6); Thioredoxin-related transmembrane protein 1 precursor (TMX1); Glypican-1 precursor (GPC1); Septin-4 isoform X1 (B4E0R1); CD177 antigen precursor (CD177); Cytoplasmic dynein 1 heavy chain 1 (DYHC1); Olfactomedin-4 precursor (OLFM4); Sorbitol dehydrogenase (DHSO); Laminin subunit beta-2 precursor (LAMB2); Myosin-9 (MYH9); Annexin A4 (ANXA4); Histone H1.3 (H13); Platelet-activating factor acetylhydrolase isoform X1 (PAFA); Glycerol kinase 2 (GLPK2); Annexin A1 (ANXA1); Integrin alpha-M isoform 1 precursor (ITAM); Peroxiredoxin-2 (PRDX2); Neprilysin isoform X1 (NEP); Dynein heavy chain 7, axonemal (DYH7); Glutamate carboxypeptidase 2 isoform 1 (FOLH1); Myeloperoxidase precursor (PERM); Annexin A5 (ANXA5); Ectonucleotide pyrophosphatase/phosphodiesterase family member 3 (ENPP3); Nucleoporin p58/p45 isoform a (NUP58); Annexin A2 isoform 2 (ANXA2); Dynein heavy chain 17, axonemal (DYH17); Voltage-dependent anion-selective channel protein 1 isoform X3 (VDAC1); Mitochondrial inner membrane protein isoform 2 (MIC60); Annexin A3 (ANXA3); Myosin light polypeptide 6 isoform 1 (MYL6); Lysosome-associated membrane glycoprotein 2 isoform C precursor (LAMP2); A-kinase anchor protein 3 (AKAP3); Aminopeptidase N isoform X1 (AMPN); Galectin-3-binding protein precursor (LG3BP); Carcinoembryonic antigen-related cell adhesion molecule 1 isoform 1 precursor (CEAM1); Nardilysin isoform a (NRDC); Peroxiredoxin-1 (PRDX1); Protein sel-1 homolog 1 isoform 1 precursor (SE1L1); Clathrin heavy chain 1 isoform 2 (A0A087WVQ6); Nuclear pore membrane glycoprotein 210 precursor (PO210); L-xylulose reductase isoform 2 (DCXR); A-kinase anchor protein 4 isoform 2 (AKAP4); Pyruvate kinase PKM isoform b (KPYM); Prostate-specific antigen isoform 1 preproprotein (KLK3); Mitochondrial carrier homolog 2 (MTCH2); Stress-70 protein, mitochondrial precursor (GRP75); Tubulin beta-4B chain (TBB4B); Prostatic acid phosphatase isoform TM-PAP precursor (PPAP); Lactotransferrin isoform 1 precursor (TRFL); Tubulin alpha-3C/D chain (TBA3C); Actin, cytoplasmic 2 (ACTG); Nuclear pore membrane glycoprotein 210-like isoform 1 precursor (P210L); Presequence protease, mitochondrial isoform 2 precursor (PREP); T-complex protein 1 subunit zeta isoform a (TCPZ); Protein disulfide-isomerase A4 precursor (PDIA4); T-complex protein 1 subunit theta isoform 1 (TCPQ); Short-chain specific acyl-coa dehydrogenase, mitochondrial precursor (ACADS); Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial precursor (ODP2); Protein disulfide-isomerase precursor (PDIA1); Very long-chain specific acyl-coa dehydrogenase, mitochondrial isoform 3 (ACADV) | Idiopathic [161] |
| Actin gamma enteric smooth muscle (ACTG2); Actin alpha skeletal muscle (ACTA1); Sperm protein associated with the nucleus on the X chromosome B F (SPANXB1); Cathelicidin antimicrobial peptide (CAMP); POTE ankyrin domain family member E (POTEE); Phospholipid hydroperoxide glutathione peroxidase mitochondrial (GPX4); Outer dense fiber protein 1 (ODF1); Clusterin (CLU); 78 kDa glucose regulated protein (HSPA5); Heat shock related 70 kDa protein 2 (HSPA2); Prolactin inducible protein (PIP); Glutathione S transferase Mu 3 (GSTM3); Tubulin beta 6 chain (TUBB6); Tubulin alpha 8 chain (TUBA8); Lactotransferrin (LTF); Triose phosphate isomerase (TPI1); A kinase anchor protein 4 (AKAP4); Glyceraldehyde 3 phosphate dehydrogenase, testis specific (GAPDHS); ATP synthase subunit beta mitochondrial (ATP5B); A kinase anchor protein 3 (AKAP3); Ras related protein Rab 2A (RAB2A); Tubulin alpha 3C D chain (TUBA3C); Ropporin 1B (ROPN1B); Histone H2B type 1 A (HIST1H2BA); Fructose bisphosphatealdolase A (ALDOA); Elongation factor 1 alpha 2 (EEF1A2); Acrosin binding protein (ACRBP); Elongation factor 1 gamma (EEF1G); Histone H2A type 1 A (HIST1H2AA); Dynein light chain 1 cytoplasmic (DYNLL1); Epididymal sperm binding protein 1 (ELSPBP1); Acrosomal protein SP 10 (ACRV1); Actin alpha cardiac muscle 1 (ACTC1) | glyceraldehyde-3-phosphate dehydrogenase, testis-specific (G3PT); Keratin type I cytoskeletal 10 (KRT10); Heat shock 70 kDa protein 1 like (HSPA1L); Tubulin beta 2C chain (TUBB2C); Heat shock cognate 71 kDa protein (HSPA8); 60S acidic ribosomal protein P2 (RPLP2); Creatine kinase B type (CKB); Prostate specific antigen (KLK3); Ubiquitin (RPS27A); Elongation factor 1 alpha 1 (EEF1A1); Tubulin beta 4 chain (TUBB4); Tubulin alpha 3E chain (TUBA3E); Keratin type II cytoskeletal 1 (KRT1); Hsc70 interacting protein (ST13); Heat shock 70 kDa protein 6 (HSPA6); Keratin type I cytoskeletal 9 (KRT9); Prostatic acid phosphatase (ACPP); Sorbitol dehydrogenase (SORD); Heat shock 70 kDa protein 1A 1B (HSPA1A); Ras related protein Rab 2B (RAB2B); Putative heat shock protein HSP 90 beta 3 (HSP90AB3P); Putative heat shock protein HSP 90 beta 4 (HSP90AB4P); Endoplasmin (HSP90B1); Beta 2 microglobulin (B2M); Putative heat shock protein HSP 90 alpha A2 (HSP90AA2); POTE ankyrin domain family member F (POTEF); Putative heat shock protein HSP 90 beta 2 (HSP90AB2P); 14 3 3 protein zeta delta (YWHAZ); Actin cytoplasmic 1 (ACTB); Heat shock protein beta 1 (HSPB1); Heat shock protein HSP 90 alpha (HSP90AA1); Beta actin like protein 2 (ACTBL2); Tubulin beta 8 chain (TUBB8); Heat shock protein HSP 90 beta (HSP90AB1); Tubulin beta 3 chain (TUBB3); Neutrophil defensin 1 (DEFA1); Putative elongation factor 1 alpha like 3 (EEF1AL3); Tubulin beta chain (TUBB); Tubulin alpha 4A chain (TUBA4A); Phospholipase A2 membrane associated (PLA2G2A); Myosin regulatory light chain 12A (MYL12A); Rho GDP dissociation inhibitor 1 (ARHGDIA); Tubulin alpha 1B chain (TUBA1B); Clusterin precursor (CLUpre);Dihydrolipoamide dehydrogenase (DLD); precursor (DLDpre); Fumarate hydratase precursor (FHpre); Heat shock-related 70 kDa protein 2 (HSPA2); Inositol-1(or 4); -monophosphatase (IMPA1); 3-mercapto-pyruvate sulfurtransferase/Delta 3,5-delta 2,4-dienoyl-CoA isomerase precursor (MPST/ ECH1pre); Proteosome beta 3 subunit human (PSMB3); Semenogelin I protein precursor (SEMG1pre); Testis-expressed sequence 12 protein (TEX12) | Astheozoopsermic [156,157] |
| Molecular Functions | FDR | Related Genes | |
|---|---|---|---|
| Fertility | Glucose metabolism | <0.001 | ENO1, PGK2, PRKACA, NUP35, TPI1, GAPDHS, ALDOA |
| TCA cycle | <0.05 | LDHC, ACO2, NDUFS1, OGDH, UQCRC2, IDH3B, MPC2, ACAD9 | |
| Infertility | Axon guidance | <0.001 | TUBA1C, TUBA3C, TUBB4B, LAMC1, MMP9, GPC1, MYH9, MYL6, A0A087WVQ6, HSPA8, RPLP2, RPS27A, TUBA3E, ACTB, HSP90AA1, TUBB8, HSP90AB1, TUBB3, TUBA4A, MYL12A, TUBA1B, PSMB3, KRT10, KRT1, KRT9, ACTB |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, Y.-J.; Pang, M.-G. Mitochondrial Functionality in Male Fertility: From Spermatogenesis to Fertilization. Antioxidants 2021, 10, 98. https://doi.org/10.3390/antiox10010098
Park Y-J, Pang M-G. Mitochondrial Functionality in Male Fertility: From Spermatogenesis to Fertilization. Antioxidants. 2021; 10(1):98. https://doi.org/10.3390/antiox10010098
Chicago/Turabian StylePark, Yoo-Jin, and Myung-Geol Pang. 2021. "Mitochondrial Functionality in Male Fertility: From Spermatogenesis to Fertilization" Antioxidants 10, no. 1: 98. https://doi.org/10.3390/antiox10010098
APA StylePark, Y. -J., & Pang, M. -G. (2021). Mitochondrial Functionality in Male Fertility: From Spermatogenesis to Fertilization. Antioxidants, 10(1), 98. https://doi.org/10.3390/antiox10010098