Human Paraoxonase-2 (PON2): Protein Functions and Modulation
Abstract
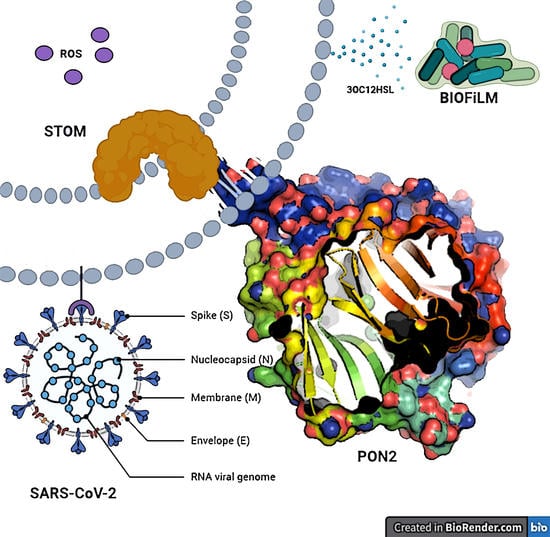
:1. Introduction
2. PON2 Structure and Function
2.1. Gene and Localization
2.2. PON2 Model
2.3. PON2 Activities
2.4. PON2 Isoforms
2.5. PON2 SNPs
2.6. PON2 Protein–Protein Interactions
3. Post-Translational Regulation of PON2
3.1. Regulation of mRNA Expression
3.2. PON2 Post-Translational Modifications
4. PON2 and the Innate Immunity
4.1. Role of Lactonase Activity
4.2. Bacteria Protect Itself from PON2 by Inducing a Post-Translational Modification (PTM)
4.3. PON2 Mediates 3OC12-HSL Biological Effects
5. PON2 and the Antioxidant Activity
5.1. Mechanism of Protection in Mitochondria
5.2. Mechanisms of Protection in ER
6. PON2 Role in Diseases
6.1. PON2 and Atherosclerosis
6.2. PON2 and Cancer
6.3. PON2 and Insulin Sensitivity
6.4. PON2 and Neurodegeneration
7. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Altenhöfer, S.; Witte, I.; Teiber, J.F.; Wilgenbus, P.; Pautz, A.; Li, H.; Daiber, A.; Witan, H.; Clement, A.M.; Förstermann, U.; et al. One Enzyme, Two Functions. J. Biol. Chem. 2010, 285, 24398–24403. [Google Scholar] [CrossRef] [Green Version]
- Devarajan, A.; Bourquard, N.; Hama, S.; Navab, M.; Grijalva, V.R.; Morvardi, S.; Clarke, C.F.; Vergnes, L.; Reue, K.; Teiber, J.F.; et al. Paraoxonase 2 Deficiency Alters Mitochondrial Function and Exacerbates the Development of Atherosclerosis. Antioxid. Redox Signal. 2011, 14, 341–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horke, S.; Witte, I.; Wilgenbus, P.; Krüger, M.; Strand, D.; Förstermann, U. Paraoxonase-2 Reduces Oxidative Stress in Vascular Cells and Decreases Endoplasmic Reticulum Stress–Induced Caspase Activation. Circulation 2007, 115, 2055–2064. [Google Scholar] [CrossRef]
- Witte, I.; Altenhöfer, S.; Wilgenbus, P.; Amort, J.; Clement, A.M.; Pautz, A.; Li, H.; Förstermann, U.; Horke, S. Beyond reduction of atherosclerosis: PON2 provides apoptosis resistance and stabilizes tumor cells. Cell Death Dis. 2011, 2, e112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Kennedy, D.; Shao, Z.; Wang, X.; Kamdar, A.K.; Weber, M.; Mislick, K.; Kiefer, K.; Morales, R.; Agatisa-Boyle, B.; et al. Paraoxonase 2 prevents the development of heart failure. Free Radic. Biol. Med. 2018, 121, 117–126. [Google Scholar] [CrossRef]
- Shakhparonov, M.I.M.I.; Antipova, N.V.; Shender, V.O.; Shnaider, P.V.; Arapidi, G.P.; Pestov, N.B.; Pavlyukov, M.S. Expression and Intracellular Localization of Paraoxonase 2 in Different Types of Malignancies. Acta Nat. 2018, 10, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Taler-Verčič, A.; Goličnik, M.; Bavec, A. The Structure and Function of Paraoxonase-1 and Its Comparison to Paraoxonase-2 and -3. Molecules 2020, 25, 5980. [Google Scholar] [CrossRef] [PubMed]
- Primo-Parmo, S.L.; Sorenson, R.C.; Teiber, J.; Du, B.N. The Human Serum Paraoxonase/Arylesterase Gene (PON1) Is One Member of a Multigene Family. Genomics 1996, 33, 498–507. [Google Scholar] [CrossRef]
- Chen, Y.; Bharill, S.; Altun, Z.; O’Hagan, R.; Coblitz, B.; Isacoff, E.Y.; Chalfie, M. Caenorhabditis elegans paraoxonase-like proteins control the functional expression of DEG/ENaC mechanosensory proteins. Mol. Biol. Cell 2016, 27, 1272–1285. [Google Scholar] [CrossRef]
- Mochizuki, H.; Scherer, S.W.; Xi, T.; Nickle, D.C.; Majer, M.; Huizenga, J.J.; Tsui, L.-C.; Prochazka, M. Human PON2 gene at 7q21.3: Cloning, multiple mRNA forms, and missense polymorphisms in the coding sequence. Gene 1998, 213, 149–157. [Google Scholar] [CrossRef]
- Diepgen, T.L.; Mallinckrodt, M.G.-V. Interethnic Differences in the Detoxification of Organophosphates: The Human Serum Paraoxonase Polymorphism. Arch. Toxicol. 1986, 9, 154–158. [Google Scholar] [CrossRef]
- Jaouad, L.; De Guise, C.; Berrougui, H.; Cloutier, M.; Isabelle, M.; Fülöp, T.; Payette, H.; Khalil, A. Age-related decrease in high-density lipoproteins antioxidant activity is due to an alteration in the PON1’s free sulfhydyl groups. Atherosclerosis 2006, 185, 191–200. [Google Scholar] [CrossRef]
- Leviev, I.; Negro, F.; James, R.W. Two Alleles of the Human Paraoxonase Gene Produce Different Amounts of mRNA. Arter. Thromb. Vasc. Biol. 1997, 17, 2935–2939. [Google Scholar] [CrossRef] [PubMed]
- Sierksma, A.; Van Der Gaag, M.S.; Van Tol, A.; James, R.W.; Hendriks, H.F.J. Kinetics of HDL cholesterol and paraoxonase activity in moderate alcohol consumers. Alcohol. Clin. Exp. Res. 2002, 26, 1430–1435. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.J.; Wadleigh, D.J.; Gangopadhyay, A.; Hama, S.; Grijalva, V.R.; Navab, M.; Fogelman, A.M.; Reddy, S.T. Paraoxonase-2 Is a Ubiquitously Expressed Protein with Antioxidant Properties and Is Capable of Preventing Cell-mediated Oxidative Modification of Low Density Lipoprotein. J. Biol. Chem. 2001, 276, 44444–44449. [Google Scholar] [CrossRef] [Green Version]
- Ng, C.J.; Shih, D.M.; Hama, S.Y.; Villa, N.; Navab, M.; Reddy, S.T. The paraoxonase gene family and atherosclerosis. Free Radic. Biol. Med. 2005, 38, 153–163. [Google Scholar] [CrossRef]
- Harel, M.; Aharoni, A.; Gaidukov, L.; Brumshtein, B.; Khersonsky, O.; Meged, R.; Dvir, H.; Ravelli, R.B.G.; McCarthy, A.; Toker, L.; et al. Structure and evolution of the serum paraoxonase family of detoxifying and anti-atherosclerotic enzymes. Nat. Struct. Mol. Biol. 2004, 11, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.L.; La Du, B.N. Calcium binding by human and rabbit serum paraoxonases. Structural stability and enzymatic activity. Drug Metab. Dispos. 1998, 26, 653–660. [Google Scholar]
- Stoltz, D.A.; Ozer, E.A.; Recker, T.J.; Estin, M.; Yang, X.; Shih, D.M.; Lusis, A.J.; Zabner, J. A Common Mutation in Paraoxonase-2 Results in Impaired Lactonase Activity. J. Biol. Chem. 2009, 284, 35564–35571. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Jiang, X.; Sun, D.; Han, G.; Wang, F.; Ye, M.; Wang, L.; Zou, H. Glycoproteomics Analysis of Human Liver Tissue by Combination of Multiple Enzyme Digestion and Hydrazide Chemistry. J. Proteome Res. 2009, 8, 651–661. [Google Scholar] [CrossRef]
- Hagmann, H.; Kuczkowski, A.; Ruehl, M.; Lamkemeyer, T.; Brodesser, S.; Horke, S.; Dryer, S.; Schermer, B.; Benzing, T.; Brinkkoetter, P.T. Breaking the chain at the membrane: Paraoxonase 2 counteracts lipid peroxidation at the plasma membrane. FASEB J. 2014, 28, 1769–1779. [Google Scholar] [CrossRef] [PubMed]
- Mandrich, L.; Cerreta, M.; Manco, G. An Engineered Version of Human PON2 Opens the Way to Understand the Role of Its Post-Translational Modifications in Modulating Catalytic Activity. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Carusone, T.M.; Cardiero, G.; Cerreta, M.; Mandrich, L.; Moran, O.; Porzio, E.; Catara, G.; Lacerra, G.; Manco, G. WTAP and BIRC3 are involved in the posttranscriptional mechanisms that impact on the expression and activity of the human lactonase PON2. Cell Death Dis. 2020, 11, 324–327. [Google Scholar] [CrossRef]
- Draganov, D.I.; Teiber, J.F.; Speelman, A.; Osawa, Y.; Sunahara, R.; La Du, B.N. Human paraoxonases (PON1, PON2, and PON3) are lactonases with overlapping and distinct substrate specificities. J. Lipid Res. 2005, 46, 1239–1247. [Google Scholar] [CrossRef] [Green Version]
- Teiber, J.F.; Horke, S.; Haines, D.C.; Chowdhary, P.K.; Xiao, J.; Kramer, G.L.; Haley, R.W.; Draganov, D.I. Dominant Role of Paraoxonases in Inactivation of the Pseudomonas aeruginosa Quorum-Sensing Signal N-(3-Oxododecanoyl)-l-Homoserine Lactone. Infect. Immun. 2008, 76, 2512–2519. [Google Scholar] [CrossRef] [Green Version]
- Sameshima, E.; Tabata, Y.; Hayashi, A.; Lida, K.; Mitsuyama, M.; Kanai, S.; Saito, T. Paraoxonase mRNA, Nirs Splice Variant1. Submitted (FEB-2003) to the EMBL/GenBank/DDBJ Databases. Available online: https://www.uniprot.org/uniprot/Q15165 (accessed on 4 January 2021).
- Ota, T.; Suzuki, Y.; Nishikawa, T.; Otsuki, T.; Sugiyama, T.; Irie, R.; Wakamatsu, A.; Hayashi, K.; Sato, H.; Nagai, K.; et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 2003, 36, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Sorenson, R.C.; Primo-Parmo, S.L.; Camper, S.A.; La Du, B.N. The Genetic Mapping and Gene Structure of Mouse Paraoxonase/Arylesterase. Genomics 1995, 30, 431–438. [Google Scholar] [CrossRef]
- Aviram, M.; Billecke, S.; Sorenson, R.; Bisgaier, C.; Newton, R.; Rosenblat, M.; Erogul, J.; Hsu, C.; Dunlop, C.; La Du, B. Paraoxonase Active Site Required for Protection Against LDL Oxidation Involves Its Free Sulfhydryl Group and Is Different From That Required for Its Arylesterase/Paraoxonase Activities. Arter. Thromb. Vasc. Biol. 1998, 18, 1617–1624. [Google Scholar] [CrossRef] [Green Version]
- Rozenberg, O.; Aviram, M. S-Glutathionylation regulates HDL-associated paraoxonase 1 (PON1) activity. Biochem. Biophys. Res. Commun. 2006, 351, 492–498. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Fan, Z.; Huang, J.; Su, S.; Yu, Q.; Zhao, J.; Hui, R.; Yao, Z.; Shen, Y.; Qiang, B.; et al. Extensive Association Analysis Between Polymorphisms of PON Gene Cluster with Coronary Heart Disease in Chinese Han Population. Arter. Thromb. Vasc. Biol. 2003, 23, 328–334. [Google Scholar] [CrossRef] [Green Version]
- Martinelli, N.; Girelli, D.; Olivieri, O.; Stranieri, C.; Trabetti, E.; Pizzolo, F.; Friso, S.; Tenuti, I.; Cheng, S.; Grow, M.A.; et al. Interaction between smoking and PON2 Ser311Cys polymorphism as a determinant of the risk of myocardial infarction. Eur. J. Clin. Investig. 2004, 34, 14–20. [Google Scholar] [CrossRef]
- Sanghera, D.K.; Aston, C.E.; Saha, N.; Kamboh, M.I. DNA Polymorphisms in Two Paraoxonase Genes (PON1 and PON2) Are Associated with the Risk of Coronary Heart Disease. Am. J. Hum. Genet. 1998, 62, 36–44. [Google Scholar] [CrossRef] [Green Version]
- Janka, Z.; Juhász, A.; Rimanóczy, Á.; Boda, K.; Márki-Zay, J.; Kálmán, J. Codon 311 (Cys → Ser) polymorphism of paraoxonase-2 gene is associated with apolipoprotein E4 allele in both Alzheimer’s and vascular dementias. Mol. Psychiatry 2002, 7, 110–112. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Zhang, S.; Tang, M.; Liu, X.; Li, T.; Han, H.; Wang, Y.; Guo, Y.; Zhao, J.; Li, H.; et al. Possible association between Cys311Ser polymorphism of paraoxonase 2 gene and late-onset Alzheimer’s disease in Chinese. Mol. Brain Res. 2004, 120, 201–204. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Ando, F.; Niino, N.; Miki, T.; Shimokata, H. Association of polymorphisms of paraoxonase 1 and 2 genes, alone or in combination, with bone mineral density in community-dwelling Japanese. J. Hum. Genet. 2003, 48, 469–475. [Google Scholar] [CrossRef]
- Hegele, R.A. Paraoxonase genes and disease. Ann. Med. 1999, 31, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Shin, B.-S.; Oh, S.-Y.; Kim, Y.-S.; Kim, K.-W. The paraoxonase gene polymorphism in stroke patients and lipid profile. Acta Neurol. Scand. 2008, 117, 237–243. [Google Scholar] [CrossRef]
- Chen, Q.; Reis, S.E.; Kammerer, C.M.; McNamara, D.M.; Holubkov, R.; Sharaf, B.L.; Sopko, G.; Pauly, D.F.; Merz, C.N.B.; Kamboh, M.I. Association between the Severity of Angiographic Coronary Artery Disease and Paraoxonase Gene Polymorphisms in the National Heart, Lung, and Blood Institute–Sponsored Women’s Ischemia Syndrome Evaluation (WISE) Study. Am. J. Hum. Genet. 2003, 72, 13–22. [Google Scholar] [CrossRef] [Green Version]
- Wheeler, J.G.; Keavney, B.D.; Watkins, H.; Collins, R.; Danesh, J. Four paraoxonase gene polymorphisms in 11 212 cases of coronary heart disease and 12 786 controls: Meta-analysis of 43 studies. Lancet 2004, 363, 689–695. [Google Scholar] [CrossRef]
- Chen, M.-L.; Zhao, H.; Liao, N.; Xie, Z.-F. Association Between Paraoxonase 2 Ser311Cys Polymorphism and Coronary Heart Disease Risk: A Meta-Analysis. Med. Sci. Monit. 2016, 22, 3196–3201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchegiani, F.; Spazzafumo, L.; Provinciali, M.; Cardelli, M.; Olivieri, F.; Franceschi, C.; Lattanzio, F.; Antonicelli, R. Paraoxonase2 C311S polymorphism and low levels of HDL contribute to a higher mortality risk after acute myocardial infarction in elderly patients. Mol. Genet. Metab. 2009, 98, 314–318. [Google Scholar] [CrossRef]
- Słowik, A.; Wloch, D.; Szermer, P.; Wolkow, P.P.; Malecki, M.; Pera, J.; Turaj, W.; Dziedzic, T.; Klimkowicz-Mrowiec, A.; Kopec, G.; et al. Paraoxonase 2 Gene C311S Polymorphism Is Associated with a Risk of Large Vessel Disease Stroke in a Polish Population. Cerebrovasc. Dis. 2007, 23, 395–400. [Google Scholar] [CrossRef]
- Porntadavity, S.; Permpongpaiboon, T.; Sukketsiri, W. Human paraoxonase 2. EXCLI J. 2010, 9, 159–172. [Google Scholar]
- Leus, F.R.; Zwart, M.; Kastelein, J.J.; Voorbij, H.A. PON2 gene variants are associated with clinical manifestations of cardiovascular disease in familial hypercholesterolemia patients. Atherosclerosis 2001, 154, 641–649. [Google Scholar] [CrossRef]
- Erlich, P.M.; Lunetta, K.L.; Cupples, L.A.; Huyck, M.; Green, R.C.; Baldwin, C.T.; Farrer, L.A.; Auerbach, S.; Akomolafe, A.; Griffith, P.; et al. Polymorphisms in the PON gene cluster are associated with Alzheimer disease. Hum. Mol. Genet. 2006, 15, 77–85. [Google Scholar] [CrossRef]
- Xie, C.; Jin, R.; Zhao, Y.; Lin, L.; Li, L.; Chen, J.; Zhang, Y. Paraoxonase 2 gene polymorphisms and prenatal phthalates’ exposure in Chinese newborns. Environ. Res. 2015, 140, 354–359. [Google Scholar] [CrossRef]
- Baig, A.; Rehman, A.-U.; Zarina, S. Association of PON2 and PON3 polymorphism with risk of developing cataract. Saudi J. Ophthalmol. 2019, 33, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Pinizzotto, M.; Castillo, E.; Fiaux, M.; Temler, E.; Gaillard, R.C.; Ruiz, J. Paraoxonase2 polymorphisms are associated with nephropathy in Type II diabetes. Diabetology 2001, 44, 104–107. [Google Scholar] [CrossRef] [Green Version]
- Qujeq, D.; Mahrooz, A.; Alizadeh, A.; Boorank, R. Paraoxonase-2 variants potentially influence insulin resistance, beta-cell function, and their interrelationships with alanine aminotransferase in type 2 diabetes. J. Res. Med. Sci. 2018, 23, 107. [Google Scholar] [PubMed]
- Mackness, B.; McElduff, P.; Mackness, M.I. The paraoxonase-2-310 polymorphism is associated with the presence of microvascular complications in diabetes mellitus. J. Intern. Med. 2005, 258, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Jäger, S.; Cimermancic, P.; Gulbahce, N.; Johnson, J.R.; McGovern, K.E.; Clarke, S.C.; Shales, M.; Mercenne, G.; Pache, L.; Li, K.; et al. Global landscape of HIV–human protein complexes. Nature 2011, 481, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Naji, S.; Ambrus, G.; Cimermančič, P.; Reyes, J.R.; Johnson, J.R.; Filbrandt, R.; Huber, M.D.; Vesely, P.; Krogan, N.J.; Yates, J.R.; et al. Host Cell Interactome of HIV-1 Rev Includes RNA Helicases Involved in Multiple Facets of Virus Production. Mol. Cell. Proteom. 2012, 11. [Google Scholar] [CrossRef] [Green Version]
- Biorxiv. Available online: https://www.biorxiv.org/content/10.1101/2020.09.03.282103v1 (accessed on 20 December 2020).
- Biorxiv. Available online: https://www.biorxiv.org/content/10.1101/2020.08.28.272955v1 (accessed on 20 December 2020).
- Nagarajan, A.; Dogra, S.K.; Sun, L.; Gandotra, N.; Ho, T.; Cai, G.; Cline, G.; Kumar, P.; Cowles, R.A.; Wajapeyee, N. Paraoxonase 2 Facilitates Pancreatic Cancer Growth and Metastasis by Stimulating GLUT1-Mediated Glucose Transport. Mol. Cell 2017, 67, 685–701.e6. [Google Scholar] [CrossRef]
- Havugimana, P.C.; Hart, G.T.; Nepusz, T.; Yang, H.; Turinsky, A.L.; Li, Z.; Wang, P.I.; Boutz, D.R.; Fong, V.; Phanse, S.; et al. A Census of Human Soluble Protein Complexes. Cell 2012, 150, 1068–1081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faraz, M.; Herdenberg, C.; Holmlund, C.; Henriksson, R.; Hedman, H. A protein interaction network centered on leucine-rich repeats and immunoglobulin-like domains 1 (LRIG1) regulates growth factor receptors. J. Biol. Chem. 2018, 293, 3421–3435. [Google Scholar] [CrossRef] [Green Version]
- Meabon, J.S.; De Laat, R.; Ieguchi, K.; Serbzhinsky, D.; Hudson, M.P.; Huber, B.R.; Wiley, J.C.; Bothwell, M. Intracellular LINGO-1 negatively regulates Trk neurotrophin receptor signaling. Mol. Cell. Neurosci. 2016, 70, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Krüger, M.; Amort, J.; Wilgenbus, P.; Helmstädter, J.P.; Grechowa, I.; Ebert, J.; Tenzer, S.; Moergel, M.; Witte, I.; Horke, S. The anti-apoptotic PON2 protein is Wnt/β-catenin-regulated and correlates with radiotherapy resistance in OSCC patients. Oncotarget 2016, 7, 51082–51095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morresi, C.; Cianfruglia, L.; Sartini, D.; Cecati, M.; Fumarola, S.; Emanuelli, M.; Armeni, T.; Ferretti, G.; Bacchetti, T. Effect of High Glucose-Induced Oxidative Stress on Paraoxonase 2 Expression and Activity in Caco-2 Cells. Cells 2019, 8, 1616. [Google Scholar] [CrossRef] [Green Version]
- Précourt, L.-P.; Marcil, V.; Ntimbane, T.; Taha, R.; Lavoie, J.-C.; Delvin, E.; Seidman, E.G.; Beaulieu, J.-F.; Levy, E. Antioxidative properties of paraoxonase 2 in intestinal epithelial cells. Am. J. Physiol. Liver Physiol. 2012, 303, G623–G634. [Google Scholar] [CrossRef] [Green Version]
- Alkhouri, R.H. Paraoxonase Gene Expression in Pediatric Inflammatory Bowel Disease. J. Clin. Cell. Immunol. 2014, 5, 224. [Google Scholar] [CrossRef] [Green Version]
- Rosenblat, M.; Draganov, D.; Watson, C.E.; Bisgaier, C.L.; La Du, B.N.; Aviram, M. Mouse Macrophage Paraoxonase 2 Activity Is Increased Whereas Cellular Paraoxonase 3 Activity Is Decreased Under Oxidative Stress. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 468–474. [Google Scholar] [CrossRef] [Green Version]
- Horiuchi, K.; Umetani, M.; Minami, T.; Okayama, H.; Takada, S.; Yamamoto, M.; Aburatani, H.; Reid, P.C.; Housman, D.E.; Hamakubo, T.; et al. Wilms’ tumor 1-associating protein regulates G2/M transition through stabilization of cyclin A2 mRNA. Proc. Natl. Acad. Sci. USA 2006, 103, 17278–17283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horiuchi, K.; Kawamura, T.; Iwanari, H.; Ohashi, R.; Naito, M.; Kodama, T.; Hamakubo, T. Identification of Wilms’ Tumor 1-associating Protein Complex and Its Role in Alternative Splicing and the Cell Cycle. J. Biol. Chem. 2013, 288, 33292–33302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuhrman, B.; Gantman, A.; Khateeb, J.; Volkova, N.; Horke, S.; Kiyan, J.; Dumler, I.; Aviram, M. Urokinase activates macrophage PON2 gene transcription via the PI3K/ROS/MEK/SREBP-2 signalling cascade mediated by the PDGFR-β. Cardiovasc. Res. 2009, 84, 145–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quang, D.; Chen, Y.; Xie, X. DANN: A deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics 2015, 31, 761–763. [Google Scholar] [CrossRef] [Green Version]
- Kim, W.; Bennett, E.J.; Huttlin, E.L.; Guo, A.; Li, J.; Possemato, A.; Sowa, M.E.; Rad, R.; Rush, J.; Comb, M.J.; et al. Systematic and Quantitative Assessment of the Ubiquitin-Modified Proteome. Mol. Cell 2011, 44, 325–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akimov, V.; Barrio-Hernandez, I.; Hansen, S.V.F.; Hallenborg, P.; Pedersen, A.-K.; Bekker-Jensen, D.B.; Puglia, M.; Christensen, S.D.K.; Vanselow, J.T.; Nielsen, M.M.; et al. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat. Struct. Mol. Biol. 2018, 25, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, N.; Rasmussen, T.B.; Jensen, P.Ø.; Stub, C.; Hentzer, M.; Molin, S.; Ciofu, O.; Givskov, M.; Johansen, H.K.; Høiby, N. Novel Mouse Model of Chronic Pseudomonas aeruginosa Lung Infection Mimicking Cystic Fibrosis. Infect. Immun. 2005, 73, 2504–2514. [Google Scholar] [CrossRef] [Green Version]
- Nakagami, G.; Morohoshi, T.; Ikeda, T.; Ohta, Y.; Sagara, H.; Huang, L.; Nagase, T.; Sugama, J.; Sanada, H. Contribution of quorum sensing to the virulence of Pseudomonas aeruginosa in pressure ulcer infection in rats. Wound Repair Regen. 2011, 19, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Christensen, L.D.; Moser, C.; Jensen, P.Ø.; Rasmussen, T.B.; Christophersen, L.; Kjelleberg, S.; Kumar, N.; Høiby, N.; Givskov, M.; Bjarnsholt, T. Impact of Pseudomonas aeruginosa quorum sensing on biofilm persistence in an in vivo intraperitoneal foreign-body infection model. Microbiology 2007, 153, 2312–2320. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Song, Z.; Hentzer, M.; Andersen, J.B.; Molin, S.; Givskov, M.; Høiby, N. Synthetic furanones inhibit quorum-sensing and enhance bacterial clearance in Pseudomonas aeruginosa lung infection in mice. J. Antimicrob. Chemother. 2004, 53, 1054–1061. [Google Scholar] [CrossRef] [Green Version]
- Streips, U.N.; Yasbin, R.E. Modern Microbial Genetics, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 2002; p. 672. ISBN 978-0-471-46108-1. [Google Scholar]
- Horke, S.; Xiao, J.; Schütz, E.-M.; Kramer, G.L.; Wilgenbus, P.; Witte, I.; Selbach, M.; Teiber, J.F. Novel Paraoxonase 2-Dependent Mechanism Mediating the Biological Effects of the Pseudomonas aeruginosa Quorum-Sensing MoleculeN-(3-Oxo-Dodecanoyl)-l-Homoserine Lactone. Infect. Immun. 2015, 83, 3369–3380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoltz, D.A.; Ozer, E.A.; Ng, C.J.; Yu, J.M.; Reddy, S.T.; Lusis, A.J.; Bourquard, N.; Parsek, M.R.; Zabner, J.; Shih, D.M. Paraoxonase-2 deficiency enhancesPseudomonas aeruginosaquorum sensing in murine tracheal epithelia. Am. J. Physiol. Cell. Mol. Physiol. 2007, 292, L852–L860. [Google Scholar] [CrossRef] [Green Version]
- Précourt, L.-P.; Amre, D.; Denis, M.-C.; Lavoie, J.-C.; Delvin, E.; Seidman, E.; Levy, E. The three-gene paraoxonase family: Physiologic roles, actions and regulation. Atherosclerosis 2011, 214, 20–36. [Google Scholar] [CrossRef] [PubMed]
- Horke, S.; Witte, I.; Altenhöfer, S.; Wilgenbus, P.; Goldeck, M.; Förstermann, U.; Xiao, J.; Kramer, G.L.; Haines, D.C.; Chowdhary, P.K.; et al. Paraoxonase 2 is down-regulated by the Pseudomonas aeruginosa quorumsensing signal N-(3-oxododecanoyl)-L-homoserine lactone and attenuates oxidative stress induced by pyocyanin. Biochem. J. 2010, 426, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Horke, S.; Witte, I.; Wilgenbus, P.; Altenhöfer, S.; Krüger, M.; Li, H.; Förstermann, U. Protective effect of paraoxonase-2 against endoplasmic reticulum stress-induced apoptosis is lost upon disturbance of calcium homoeostasis. Biochem. J. 2008, 416, 395–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schweikert, E.-M.; Amort, J.; Wilgenbus, P.; Förstermann, U.; Teiber, J.F.; Horke, S. Paraoxonases-2 and -3 Are Important Defense Enzymes againstPseudomonas aeruginosaVirulence Factors due to Their Anti-Oxidative and Anti-Inflammatory Properties. J. Lipids 2012, 2012, 352857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teplitski, M.; Mathesius, U.; Rumbaugh, K.P. Perception and Degradation ofN-Acyl Homoserine Lactone Quorum Sensing Signals by Mammalian and Plant Cells. Chem. Rev. 2011, 111, 100–116. [Google Scholar] [CrossRef]
- Cadenas, E.; Davies, K.J. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic. Biol. Med. 2000, 29, 222–230. [Google Scholar] [CrossRef]
- Madamanchi, N.R.; Runge, M.S. Mitochondrial Dysfunction in Atherosclerosis. Circ. Res. 2007, 100, 460–473. [Google Scholar] [CrossRef] [Green Version]
- Zimmermann, S.; Wagner, C.; Müller, W.; Brenner-Weiss, G.; Hug, F.; Prior, B.; Obst, U.; Hänsch, G.M. Induction of Neutrophil Chemotaxis by the Quorum-Sensing Molecule N-(3-Oxododecanoyl)-l-Homoserine Lactone. Infect. Immun. 2006, 74, 5687–5692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tateda, K.; Ishii, Y.; Horikawa, M.; Matsumoto, T.; Miyairi, S.; Pechere, J.C.; Standiford, T.J.; Ishiguro, M.; Yamaguchi, K. The Pseudomonas aeruginosa Autoinducer N-3-Oxododecanoyl Homoserine Lactone Accelerates Apoptosis in Macrophages and Neutrophils. Infect. Immun. 2003, 71, 5785–5793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.B.; Xia, Y.-R.; Romanoski, C.E.; Lee, S.; Meng, Y.; Shi, Y.-S.; Bourquard, N.; Gong, K.W.; Port, Z.; Grijalva, V.; et al. Paraoxonase-2 Modulates Stress Response of Endothelial Cells to Oxidized Phospholipids and a Bacterial Quorum–Sensing Molecule. Arter. Thromb. Vasc. Biol. 2011, 31, 2624–2633. [Google Scholar] [CrossRef] [Green Version]
- Schwarzer, C.; Fu, Z.; Morita, T.; Whitt, A.G.; Neely, A.M.; Liu, Y.; Machen, T.E.; Sun, B.; Xiao, Z.; Wang, R.; et al. Paraoxonase 2 Serves a Proapopotic Function in Mouse and Human Cells in Response to the Pseudomonas aeruginosa Quorum-sensing Molecule N-(3-Oxododecanoyl)-homoserine Lactone. J. Biol. Chem. 2015, 290, 7247–7258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, S.; Luo, Y.; He, B.; Liu, J.; Qian, X.; Ni, Y.; Zhao, R. Paraoxonase 2 modulates a proapoptotic function in LS174T cells in response to quorum sensing molecule N-(3-oxododecanoyl)-L-homoserine lactone. Sci. Rep. 2016, 6, 28778. [Google Scholar] [CrossRef]
- Tao, S.; Niu, L.; Cai, L.; Geng, Y.; Hua, C.; Ni, Y.D.; Zhao, R. N -(3-oxododecanoyl)- l -homoserine lactone modulates mitochondrial function and suppresses proliferation in intestinal goblet cells. Life Sci. 2018, 201, 81–88. [Google Scholar] [CrossRef]
- Zhao, G.; Neely, A.M.; Schwarzer, C.; Lu, H.; Whitt, A.G.; Stivers, N.S.; Burlison, J.A.; White, C.; Machen, T.E.; Li, C. N-(3-oxo-acyl) homoserine lactone inhibits tumor growth independent of Bcl-2 proteins. Oncotarget 2016, 7, 5924–5942. [Google Scholar] [CrossRef]
- Madamanchi, N.R.; Vendrov, A.; Runge, M.S. Oxidative Stress and Vascular Disease. Arter. Thromb. Vasc. Biol. 2005, 25, 29–38. [Google Scholar] [CrossRef] [Green Version]
- Aliev, G.; Priyadarshini, M.; Reddy, V.P.; Grieg, N.; Kaminsky, Y.; Cacabelos, R.; Ashraf, G.M.; Jabir, N.; Kamal, M.A.; Nikolenko, V.N.; et al. Oxidative Stress Mediated Mitochondrial and Vascular Lesions as Markers in the Pathogenesis of Alzheimer Disease. Curr. Med. Chem. 2014, 21, 2208–2217. [Google Scholar] [CrossRef]
- Gaki, G.S.; Papavassiliou, A.G. Oxidative Stress-Induced Signaling Pathways Implicated in the Pathogenesis of Parkinson’s Disease. Neuromolecular Med. 2014, 16, 217–230. [Google Scholar] [CrossRef]
- Landriscina, M.; Maddalena, F.; Laudiero, G.; Esposito, F. Adaptation to Oxidative Stress, Chemoresistance, and Cell Survival. Antioxid. Redox Signal. 2009, 11, 2701–2716. [Google Scholar] [CrossRef] [PubMed]
- Higgins, G.C.; Beart, P.M.; Shin, Y.S.; Chen, M.J.; Cheung, N.S.; Nagley, P. Oxidative Stress: Emerging Mitochondrial and Cellular Themes and Variations in Neuronal Injury. J. Alzheimers Dis. 2010, 20, S453–S473. [Google Scholar] [CrossRef] [Green Version]
- Ng, C.J.; Bourquard, N.; Grijalva, V.; Hama, S.; Shih, D.M.; Navab, M.; Fogelman, A.M.; Lusis, A.J.; Young, S.; Reddy, S.T. Paraoxonase-2 Deficiency Aggravates Atherosclerosis in Mice Despite Lower Apolipoprotein-B-containing Lipoproteins. J. Biol. Chem. 2006, 281, 29491–29500. [Google Scholar] [CrossRef] [Green Version]
- Levy, E.; Trudel, K.; Bendayan, M.; Seidman, E.; Delvin, E.; Elchebly, M.; Lavoie, J.-C.; Precourt, L.-P.; Amre, D.; Sinnett, D. Biological role, protein expression, subcellular localization, and oxidative stress response of paraoxonase 2 in the intestine of humans and rats. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 293, G1252–G1261. [Google Scholar] [CrossRef] [Green Version]
- Precourt, L.-P.; Seidman, E.; Delvin, E.; Amre, D.; Deslandres, C.; Dominguez, M.; Sinnett, D.; Levy, E. Comparative expression analysis reveals differences in the regulation of intestinal paraoxonase family members. Int. J. Biochem. Cell Biol. 2009, 41, 1628–1637. [Google Scholar] [CrossRef]
- Rosenblat, M.; Coleman, R.; Reddy, S.T.; Aviram, M. Paraoxonase 2 attenuates macrophage triglyceride accumulation via inhibition of diacylglycerol acyltransferase 1. J. Lipid Res. 2009, 50, 870–879. [Google Scholar] [CrossRef] [Green Version]
- Meilin, E.; Aviram, M.; Hayek, T. Paraoxonase 2 (PON2) decreases high glucose-induced macrophage triglycerides (TG) accumulation, via inhibition of NADPH-oxidase and DGAT1 activity: Studies in PON2-deficient mice. Atherosclerosis 2010, 208, 390–395. [Google Scholar] [CrossRef] [PubMed]
- Costa, L.G.; De Laat, R.; Dao, K.; Pellacani, C.; Cole, T.B.; Furlong, C.E. Paraoxonase-2 (PON2) in brain and its potential role in neuroprotection. Neurotoxicology 2014, 43, 3–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sulaiman, D.; Li, J.; Devarajan, A.; Cunningham, C.M.; Li, M.; Fishbein, G.A.; Fogelman, A.M.; Eghbali, M.; Reddy, S.T. Paraoxonase 2 protects against acute myocardial ischemia-reperfusion injury by modulating mitochondrial function and oxidative stress via the PI3K/Akt/GSK-3β RISK pathway. J. Mol. Cell. Cardiol. 2019, 129, 154–164. [Google Scholar] [CrossRef]
- Ohnishi, T.; Trumpower, B. Differential effects of antimycin on ubisemiquinone bound in different environments in isolated succinate. cytochrome c reductase complex. J. Biol. Chem. 1980, 255, 3278–3284. [Google Scholar] [CrossRef]
- Linnane, A.W.; Eastwood, H. Cellular Redox Regulation and Prooxidant Signaling Systems: A New Perspective on the Free Radical Theory of Aging. Ann. N. Y. Acad. Sci. 2006, 1067, 47–55. [Google Scholar] [CrossRef]
- Turrens, J.F.; Alexandre, A.; Lehninger, A.L. Ubisemiquinone is the electron donor for superoxide formation by complex III of heart mitochondria. Arch. Biochem. Biophys. 1985, 237, 408–414. [Google Scholar] [CrossRef]
- Bourquard, N.; Ng, C.J.; Reddy, S.T. Impaired hepatic insulin signalling in PON2-deficient mice: A novel role for the PON2/apoE axis on the macrophage inflammatory response. Biochem. J. 2011, 436, 91–100. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, H.; Haze, K.; Yanagi, H.; Yura, T.; Mori, K. Identification of the cis-Acting Endoplasmic Reticulum Stress Response Element Responsible for Transcriptional Induction of Mammalian Glucose-regulated Proteins. J. Biol. Chem. 1998, 273, 33741–33749. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Kaufman, R.J. The unfolded protein response: A stress signaling pathway critical for health and disease. Neurology 2005, 66, S102–S109. [Google Scholar] [CrossRef] [PubMed]
- Porter, A.G.; Ja, R.U.; Nicke, È. Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 1999, 6, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Marsillach, J.; Mackness, B.; Mackness, M.; Riu, F.; Beltrán, R.; Joven, J.; Camps, J. Immunohistochemical analysis of paraoxonases-1, 2, and 3 expression in normal mouse tissues. Free Radic. Biol. Med. 2008, 45, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.J.; Hama, S.Y.; Bourquard, N.; Navab, M.; Reddy, S.T. Adenovirus mediated expression of human paraoxonase 2 protects against the development of atherosclerosis in apolipoprotein E-deficient mice. Mol. Genet. Metab. 2006, 89, 368–373. [Google Scholar] [CrossRef]
- Lin, J.H.; Li, H.; Yasumura, D.; Cohen, H.R.; Zhang, C.; Panning, B.; Shokat, K.M.; Lavail, M.M.; Walter, P. IRE1 Signaling Affects Cell Fate During the Unfolded Protein Response. Science 2007, 318, 944–949. [Google Scholar] [CrossRef] [Green Version]
- Urano, F.; Wang, X.; Bertolotti, A.; Zhang, Y.; Chung, P.; Harding, H.P.; Ron, D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 2000, 287, 664–666. [Google Scholar] [CrossRef] [Green Version]
- Calfon, M.; Zeng, H.; Urano, F.; Till, J.H.; Hubbard, S.R.; Harding, H.P.; Clark, G.S.; Ron, D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 2002, 415, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Sato, T.; Matsui, T.; Sato, M.; Okada, T.; Yoshida, H.; Harada, A.; Mori, K. Transcriptional Induction of Mammalian ER Quality Control Proteins Is Mediated by Single or Combined Action of ATF6α and XBP1. Dev. Cell 2007, 13, 365–376. [Google Scholar] [CrossRef] [Green Version]
- Lee, A.-H.; Iwakoshi, N.N.; Glimcher, L.H. XBP-1 Regulates a Subset of Endoplasmic Reticulum Resident Chaperone Genes in the Unfolded Protein Response. Mol. Cell. Biol. 2003, 23, 7448–7459. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.; Tirasophon, W.; Shen, X.; Michalak, M.; Prywes, R.; Okada, T.; Yoshida, H.; Mori, K.; Kaufman, R.J. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002, 16, 452–466. [Google Scholar] [CrossRef] [Green Version]
- Rothe, M.; Sarma, V.; Dixit, V.M.; Goeddel, D.V. TRAF2-mediated activation of NF-kappa B by TNF receptor 2 and CD40. Science 1995, 269, 1424–1427. [Google Scholar] [CrossRef]
- Malhotra, J.D.; Kaufman, R.J. Endoplasmic Reticulum Stress and Oxidative Stress: A Vicious Cycle or a Double-Edged Sword? Antioxid. Redox Signal. 2007, 9, 2277–2294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimizu, Y.; Hendershot, L.M. Oxidative Folding: Cellular Strategies for Dealing with the Resultant Equimolar Production of Reactive Oxygen Species. Antioxid. Redox Signal. 2009, 11, 2317–2331. [Google Scholar] [CrossRef]
- Shen, H.-M.; Liu, Z.-G. JNK signaling pathway is a key modulator in cell death mediated by reactive oxygen and nitrogen species. Free Radic. Biol. Med. 2006, 40, 928–939. [Google Scholar] [CrossRef]
- Hu, H.; Tian, M.; Ding, C.; Yu, S. The C/EBP Homologous Protein (CHOP) Transcription Factor Functions in Endoplasmic Reticulum Stress-Induced Apoptosis and Microbial Infection. Front. Immunol. 2019, 9, 3083. [Google Scholar] [CrossRef] [Green Version]
- Oyadomari, S.; Mori, M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2003, 11, 381–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.; Yang, J.; Feng, B.; Lu, W.; Zhao, C.; Li, L. Mendelian randomization analysis identified genes pleiotropically associated with the risk and prognosis of COVID-19. J. Infect. 2021, 82, 126–132. [Google Scholar] [CrossRef]
- Ebert, J.; Wilgenbus, P.; Teiber, J.F.; Jurk, K.; Schwierczek, K.; Döhrmann, M.; Xia, N.; Li, H.; Spiecker, L.; Ruf, W.; et al. Paraoxonase-2 regulates coagulation activation through endothelial tissue factor. Blood 2018, 131, 2161–2172. [Google Scholar] [CrossRef]
- Xu, J.H.; Lu, S.J.; Wu, P.; Kong, L.C.; Ning, C.; Li, H.Y. Molecular mechanism whereby paraoxonase-2 regulates coagulation activation through endothelial tissue factor in rat haemorrhagic shock model. Int. Wound J. 2020, 17, 735–741. [Google Scholar] [CrossRef]
- Reddy, S.T.; Wadleigh, D.J.; Grijalva, V.; Ng, C.; Hama, S.; Gangopadhyay, A.; Shih, D.M.; Lusis, A.J.; Navab, M.; Fogelman, A.M. Human paraoxonase-3 is an HDL-associated enzyme with biological activity similar to paraoxonase-1 protein but is not regulated by oxidized lipids. Arter. Thromb. Vasc. Biol. 2001, 21, 542–547. [Google Scholar] [CrossRef] [Green Version]
- Gotto, A.M.; Pownall, H.J.; Havel, R.J. [1] Introduction to the plasma lipoproteins. Biomembr. Part K Membr. Biog. Assembly Target. (Prokaryotes Mitochondria Chloroplasts) 1986, 128, 3–41. [Google Scholar] [CrossRef]
- Ramos, P.; Gieseg, S.P.; Schuster, B.; Esterbauer, H. Effect of temperature and phase transition on oxidation resistance of low density lipoprotein. J. Lipid Res. 1996, 36, 2113–2128. [Google Scholar] [CrossRef]
- Mertens, A.; Holvoet, P. Oxidized LDL and HDL: Antagonists in atherothrombosis. FASEB J. 2001, 15, 2073–2084. [Google Scholar] [CrossRef] [Green Version]
- Taleb, S. Inflammation in atherosclerosis. Arch. Cardiovasc. Dis. 2016, 109, 708–715. [Google Scholar] [CrossRef]
- Steinberg, D. Low Density Lipoprotein Oxidation and Its Pathobiological Significance. J. Biol. Chem. 1997, 272, 20963–20966. [Google Scholar] [CrossRef] [Green Version]
- Mantovani, A.; Sica, A. Macrophages, innate immunity and cancer: Balance, tolerance, and diversity. Curr. Opin. Immunol. 2010, 22, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Moloney, J.N.; Cotter, T.G. ROS signalling in the biology of cancer. Semin. Cell Dev. Biol. 2018, 80, 50–64. [Google Scholar] [CrossRef] [PubMed]
- Panieri, E.; Santoro, M. ROS homeostasis and metabolism: A dangerous liason in cancer cells. Cell Death Dis. 2016, 7, e2253. [Google Scholar] [CrossRef]
- Liou, G.Y.; Storz, P. Reactive oxygen species in cancer. Free Radic. Res. 2010, 44, 479–496. [Google Scholar] [CrossRef] [Green Version]
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic. Biol. Med. 2010, 49, 1603–1616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janssen-Heininger, Y.M.W.; Mossman, B.T.; Heintz, N.H.; Forman, H.J.; Kalyanaraman, B.; Finkel, T.; Stamler, J.S.; Rhee, S.G.; Van Der Vliet, A. Redox-based regulation of signal transduction: Principles, pitfalls, and promises. Free Radic. Biol. Med. 2008, 45, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Benhar, M.; Engelberg, D.; Levitzki, A. ROS, stress-activated kinases and stress signaling in cancer. EMBO Rep. 2002, 3, 420–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Witte, I.; Foerstermann, U.; Devarajan, A.; Reddy, S.T.; Horke, S. Protectors or Traitors: The Roles of PON2 and PON3 in Atherosclerosis and Cancer. J. Lipids 2012, 2012, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Bacchetti, T.; Ferretti, G.; Sahebkar, A. The role of paraoxonase in cancer. Semin. Cancer Biol. 2019, 56, 72–86. [Google Scholar] [CrossRef]
- Li, Y.; Li, Y.; Tang, R.; Xu, H.; Qiu, M.; Chen, Q.; Chen, J.; Fu, Z.; Ying, K.; Xie, Y.; et al. Discovery and analysis of hepatocellular carcinoma genes using cDNA microarrays. J. Cancer Res. Clin. Oncol. 2002, 128, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Ribarska, T.; Ingenwerth, M.; Goering, W.; Engers, R.; Schulz, W.A. Epigenetic inactivation of the placentally imprinted tumor suppressor gene TFPI2 in prostate carcinoma. Cancer Genom. Proteom. 2010, 7, 51–60. [Google Scholar]
- Bacchetti, T.; Salvolini, E.; Pompei, V.; Campagna, R.; Molinelli, E.; Brisigotti, V.; Togni, L.; Lucarini, G.; Sartini, D.; Campanati, A.; et al. Paraoxonase-2: A potential biomarker for skin cancer aggressiveness. Eur. J. Clin. Investig. 2020, e13452. [Google Scholar] [CrossRef]
- Wang, X.; Xu, G.; Zhang, J.; Wang, S.; Ji, M.; Mo, L.; Zhu, M.; Li, J.; Zhou, G.; Lu, J.; et al. The clinical and prognostic significance of paraoxonase-2 in gastric cancer patients: Immunohistochemical analysis. Hum. Cell 2019, 32, 487–494. [Google Scholar] [CrossRef]
- Wang, R.; Li, J.; Zhao, Y.; Li, Y.; Yin, L. Investigating the therapeutic potential and mechanism of curcumin in breast cancer based on RNA sequencing and bioinformatics analysis. Breast Cancer 2017, 25, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Ross, M.E.; Zhou, X.; Song, G.; Shurtleff, S.A.; Girtman, K.; Williams, W.K.; Liu, H.-C.; Mahfouz, R.; Raimondi, S.; Lenny, N.; et al. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood 2003, 102, 2951–2959. [Google Scholar] [CrossRef] [Green Version]
- Kang, H.; Chen, I.-M.; Wilson, C.S.; Bedrick, E.J.; Harvey, R.C.; Atlas, S.R.; Devidas, M.; Mullighan, C.G.; Wang, X.; Murphy, M.; et al. Gene expression classifiers for relapse-free survival and minimal residual disease improve risk classification and outcome prediction in pediatric B-precursor acute lymphoblastic leukemia. Blood 2010, 115, 1394–1405. [Google Scholar] [CrossRef]
- Frank, O.; Brors, B.; Fabarius, A.; Li, L.; Haak, M.; Merk, S.; Schwindel, U.; Zheng, C.; Müller, M.C.; Gretz, N.; et al. Gene expression signature of primary imatinib-resistant chronic myeloid leukemia patients. Leukemia 2006, 20, 1400–1407. [Google Scholar] [CrossRef] [Green Version]
- Pise-Masison, C.A.; Radonovich, M.; Mahieux, R.; Chatterjee, P.; Whiteford, C.; Duvall, J.; Guillerm, C.; Gessain, A.; Brady, J.N. Transcription profile of cells infected with human T-cell leukemia virus type I compared with activated lymphocytes. Cancer Res. 2002, 62, 3562–3571. [Google Scholar]
- Bacchetti, T.; Sartini, D.; Pozzi, V.; Cacciamani, T.; Ferretti, G.; Emanuelli, M. Exploring the role of Paraoxonase-2 in bladder cancer: Analyses performed on tissue samples, urines and cell culturess. Oncotarget 2017, 8, 28785–28795. [Google Scholar] [CrossRef] [Green Version]
- Tseng, J.-H.; Chen, C.-Y.; Chen, P.-C.; Hsiao, S.-H.; Fan, C.-C.; Liang, Y.-C.; Chen, C.-P. Valproic acid inhibits glioblastoma multiforme cell growth via paraoxonase 2 expression. Oncotarget 2017, 8, 14666–14679. [Google Scholar] [CrossRef] [Green Version]
- Shiner, M.; Fuhrman, B.; Aviram, M. Paraoxonase 2 (PON2) expression is upregulated via a reduced-nicotinamide-adenine-dinucleotide-phosphate (NADPH)-oxidase-dependent mechanism during monocytes differentiation into macrophages. Free Radic. Biol. Med. 2004, 37, 2052–2063. [Google Scholar] [CrossRef] [PubMed]
- Azizi, F.; Rahmani, M.; Raiszadeh, F.; Solati, M.; Navab, M. Association of lipids, lipoproteins, apolipoproteins and paraoxonase enzyme activity with premature coronary artery disease. Coron. Artery Dis. 2002, 13, 9–16. [Google Scholar] [CrossRef]
- Schweikert, E.-M.; Devarajan, A.; Witte, I.; Wilgenbus, P.; Amort, J.; Förstermann, U.; Shabazian, A.; Grijalva, V.; Shih, D.M.; Farias-Eisner, R.; et al. PON3 is upregulated in cancer tissues and protects against mitochondrial superoxide-mediated cell death. Cell Death Differ. 2012, 19, 1549–1560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krüger, M.; Pabst, A.; Al-Nawas, B.; Horke, S.; Moergel, M. Paraoxonase-2 (PON2) protects oral squamous cell cancer cells against irradiation-induced apoptosis. J. Cancer Res. Clin. Oncol. 2015, 141, 1757–1766. [Google Scholar] [CrossRef]
- Devarajan, A.; Su, F.; Grijalva, V.; Yalamanchi, M.; Yalamanchi, A.; Gao, F.; Trost, H.; Nwokedi, J.; Farias-Eisner, G.; Farias-Eisner, R.; et al. Paraoxonase 2 overexpression inhibits tumor development in a mouse model of ovarian cancer. Cell Death Dis. 2018, 9, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klover, P.J.; Zimmers, T.A.; Koniaris, L.G.; Mooney, R.A. Chronic Exposure to Interleukin-6 Causes Hepatic Insulin Resistance in Mice. Diabetes 2003, 52, 2784–2789. [Google Scholar] [CrossRef] [Green Version]
- Shoelson, S.E.; Lee, J.; Goldfine, A.B. Inflammation and insulin resistance. J. Clin. Investig. 2006, 116, 1793–1801. [Google Scholar] [CrossRef] [PubMed]
- Cai, D.; Yuan, M.; Frantz, D.F.; Melendez, P.A.; Hansen, L.; Lee, J.; Shoelson, S.E. Local and systemic insulin resistance resulting from hepatic activation of IKK-β and NF-κB. Nat. Med. 2005, 11, 183–190. [Google Scholar] [CrossRef]
- Song, B.; Scheuner, D.; Ron, D.; Pennathur, S.; Kaufman, R.J. Chop deletion reduces oxidative stress, improves β cell function, and promotes cell survival in multiple mouse models of diabetes. J. Clin. Investig. 2008, 118, 3378–3389. [Google Scholar] [CrossRef] [Green Version]
- Giordano, G.; Cole, T.B.; Furlong, C.E.; Costa, L.G. Paraoxonase 2 (PON2) in the mouse central nervous system: A neuroprotective role? Toxicol. Appl. Pharmacol. 2011, 256, 369–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mackness, B.; Beltran-Debon, R.; Aragones, G.; Joven, J.; Camps, J.; Mackness, M. Human tissue distribution of paraoxonases 1 and 2 mRNA. IUBMB Life 2010, 62, 480–482. [Google Scholar] [CrossRef]
- Costa, L.G.; Tait, L.; De Laat, R.; Dao, K.; Giordano, G.; Pellacani, C.; Cole, T.B.; Furlong, C.E. Modulation of Paraoxonase 2 (PON2) in Mouse Brain by the Polyphenol Quercetin: A Mechanism of Neuroprotection? Neurochem. Res. 2013, 38, 1809–1818. [Google Scholar] [CrossRef] [Green Version]
- Shamir, R.; Hartman, C.; Karry, R.; Pavlotzky, E.; Eliakim, R.; Lachter, J.; Suissa, A.; Aviram, M. Paraoxonases (PONs) 1, 2, and 3 are expressed in human and mouse gastrointestinal tract and in Caco-2 cell line: Selective secretion of PON1 and PON2. Free Radic. Biol. Med. 2005, 39, 336–344. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.; Luo, D.; Yang, M.; Wang, Y.; Xiong, L.; Gao, L.; Liu, Y.; Liu, H. A Meta-Analysis on the Relationship of the PON Genes and Alzheimer Disease. J. Geriatr. Psychiatry Neurol. 2017, 30, 303–310. [Google Scholar] [CrossRef] [PubMed]
- LeDuc, V.; Legault, V.; Dea, D.; Poirier, J. Normalization of gene expression using SYBR green qPCR: A case for paraoxonase 1 and 2 in Alzheimer’s disease brains. J. Neurosci. Methods 2011, 200, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Mu, N.; Xu, S.C.; Chang, Q.; Rao, D.P.; Chen, J.P.; Ma, C. Study of lipids, insulin metabolism, and paraoxonase-2-311 polymorphism in patients with different subtypes of Alzheimer’s disease (translated version). East Asian Arch. Psychiatry 2013, 23, 114–119. [Google Scholar]
- Akhmedova, S.N.; Yakimovsky, A.K.; Schwartz, E.I. Paraoxonase 1 Met–Leu 54 polymorphism is associated with Parkinson’s disease. J. Neurol. Sci. 2001, 184, 179–182. [Google Scholar] [CrossRef]
- Carmine, A.; Buervenich, S.; Sydow, O.; Anvret, M.; Olson, L. Further evidence for an association of the Paraoxonase 1 (PON1) Met-54 allele with Parkinson’s disease. Mov. Disord. 2002, 17, 764–766. [Google Scholar] [CrossRef]
- Hirsch, E.C.; Graybiel, A.M.; Agid, Y.A. Melanized dopaminergic neurons are differentially susceptible to degeneration in Parkinson’s disease. Nat. Cell Biol. 1988, 334, 345–348. [Google Scholar] [CrossRef] [PubMed]
- Tanner, C.M. Parkinson Disease in TwinsAn Etiologic Study. JAMA 1999, 281, 341–346. [Google Scholar] [CrossRef]
- Polymeropoulos, M.H. Mutation in the -Synuclein Gene Identified in Families with Parkinson’s Disease. Science 1997, 276, 2045–2047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitada, T.; Asakawa, S.; Hattori, N.; Matsumine, H.; Yamamura, Y.; Minoshima, S.; Yokochi, M.; Mizuno, Y.; Shimizu, N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nat. Cell Biol. 1998, 392, 605–608. [Google Scholar] [CrossRef] [PubMed]
- Leroy, E.; Boyer, R.; Auburger, G.; Leube, B.; Ulm, G.; Mezey, E.; Harta, G.; Brownstein, M.J.; Jonnalagada, S.; Chernova, T.; et al. The ubiquitin pathway in Parkinson’s disease. Nat. Cell Biol. 1998, 395, 451–452. [Google Scholar] [CrossRef] [PubMed]
- Krüger, R.; Kuhn, W.; Müller, T.; Woitalla, D.; Graeber, M.B.; Kösel, S.; Przuntek, H.; Epplen, J.T.; Schols, L.; Riess, O. AlaSOPro mutation in the gene encoding α-synuclein in Parkinson’s disease. Nat. Genet. 1998, 18, 106–108. [Google Scholar] [CrossRef] [PubMed]
- Bonifati, V.; Rizzu, P.; Van Baren, M.J.; Schaap, O.; Breedveld, G.J.; Krieger, E.; Dekker, M.C.J.; Squitieri, F.; Ibanez, P.; Joosse, M.; et al. Mutations in the DJ-1 Gene Associated with Autosomal Recessive Early-Onset Parkinsonism. Science 2002, 299, 256–259. [Google Scholar] [CrossRef] [Green Version]
- Ewing, R.M.; Chu, P.; Elisma, F.; Li, H.; Taylor, P.; Climie, S.; McBroom-Cerajewski, L.; Robinson, M.D.; O’Connor, L.; Li, M.; et al. Large-scale mapping of human protein–protein interactions by mass spectrometry. Mol. Syst. Biol. 2007, 3, 89. [Google Scholar] [CrossRef] [PubMed]
- Parsanejad, M.; Bourquard, N.; Qu, D.; Zhang, Y.; Huang, E.; Rousseaux, M.W.C.; Aleyasin, H.; Irrcher, I.; Callaghan, S.; Vaillant, D.C.; et al. DJ-1 Interacts with and Regulates Paraoxonase-2, an Enzyme Critical for Neuronal Survival in Response to Oxidative Stress. PLoS ONE 2014, 9, e106601. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Kim, K.S.; Iyirhiaro, G.O.; Marcogliese, P.C.; Callaghan, S.M.; Qu, D.; Kim, W.J.; Slack, R.S.; Park, D.S. DJ-1 modulates the unfolded protein response and cell death via upregulation of ATF4 following ER stress. Cell Death Dis. 2019, 10, 135. [Google Scholar] [CrossRef] [Green Version]
- Holtz, W.A.; O’Malley, K.L. Parkinsonian Mimetics Induce Aspects of Unfolded Protein Response in Death of Dopaminergic Neurons. J. Biol. Chem. 2003, 278, 19367–19377. [Google Scholar] [CrossRef] [Green Version]
- Aguirre-Vidal, Y.; Morales-Montor, J.; De León, C.T.G.; Ostoa-Saloma, P.; Díaz-Zaragoza, M.; Montes, S.; Arteaga-Silva, M.; Antonio, M.-N. Protection induced by estradiol benzoate in the MPP+ rat model of Parkinson’s disease is associated with the regulation of the inflammatory cytokine profile in the nigro striatum. J. Neuroimmunol. 2020, 349, 577426. [Google Scholar] [CrossRef]
- Reichert, C.O.; Levy, D.; Bydlowski, S.P. Paraoxonase Role in Human Neurodegenerative Diseases. Antioxidants 2020, 10, 11. [Google Scholar] [CrossRef]
- Narain, P.; Padhi, A.K.; Dave, U.; Mishra, D.; Bhatia, R.; Vivekanandan, P.; Gomes, J. Identification and characterization of novel and rare susceptible variants in Indian amyotrophic lateral sclerosis patients. Neurogenetics 2019, 20, 197–208. [Google Scholar] [CrossRef]
- Obrador, E.; Salvador, R.; López-Blanch, R.; Jihad-Jebbar, A.; Vallés, S.L.; Estrela, J.M. Oxidative Stress, Neuroinflammation and Mitochondria in the Pathophysiology of Amyotrophic Lateral Sclerosis. Antioxidants 2020, 9, 901. [Google Scholar] [CrossRef]
- Herrando-Grabulosa, M.; Gaja-Capdevila, N.; Vela, J.M.; Navarro, X. Sigma 1 receptor as a therapeutic target for amyotrophic lateral sclerosis. Br. J. Pharmacol. 2020, 72, 1–17. [Google Scholar] [CrossRef]
- Saeed, M.; Siddique, N.; Hung, W.Y.; Usacheva, E.; Liu, E.; Sufit, R.L.; Heller, S.L.; Haines, J.L.; Pericak-Vance, M.; Siddique, T. Paraoxonase cluster polymorphisms are associated with sporadic ALS. Neurology 2006, 67, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Slowik, A.; Tomik, B.; Wolkow, P.P.; Partyka, D.; Turaj, W.; Malecki, M.T.; Pera, J.; Dziedzic, T.; Szczudlik, A.; Figlewicz, D.A. Paraoxonase gene polymorphisms and sporadic ALS. Neurology 2006, 67, 766–770. [Google Scholar] [CrossRef]
- Landers, J.E.; Shi, L.; Cho, T.-J.; Glass, J.D.; Shaw, P.; Leigh, P.N.; Diekstra, F.; Polak, M.; Rodriguez-Leyva, I.; Niemann, S.; et al. A common haplotype within the PON1 promoter region is associated with sporadic ALS. Amyotroph. Lateral Scler. 2008, 9, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Valdmanis, P.N.; Kabashi, E.; Dyck, A.; Hince, P.; Lee, J.; Dion, P.; D’Amour, M.; Souchon, F.; Bouchard, J.-P.; Salachas, F.; et al. Association of paraoxonase gene cluster polymorphisms with ALS in France, Quebec, and Sweden. Neurology 2008, 71, 514–520. [Google Scholar] [CrossRef]
- Ticozzi, N.; Ba, A.L.L.; Bs, P.J.K.; Glass, J.D.; Wills, A.-M.; Van Blitterswijk, M.; Bosco, D.A.; Rodriguez-Leyva, I.; Gellera, C.; Ratti, A.; et al. Paraoxonase gene mutations in amyotrophic lateral sclerosis. Ann. Neurol. 2010, 68, 102–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricci, C.; Battistini, S.; Cozzi, L.; Benigni, M.; Origone, P.; Verriello, L.; Lunetta, C.; Cereda, C.; Milani, P.; Greco, G.; et al. Lack of association of PON polymorphisms with sporadic ALS in an Italian population. Neurobiol. Aging 2011, 32, 552.e7–552.e13. [Google Scholar] [CrossRef]
- Chen, Y.; Huang, R.; Chen, K.; Song, W.; Yang, Y.; Zhao, B.; Li, J.; Shang, H.-F. Association analysis of PON polymorphisms in sporadic ALS in a Chinese population. Neurobiol. Aging 2012, 33, 2949.e1–2949.e3. [Google Scholar] [CrossRef]
- Gagliardi, S.; Abel, K.; Bianchi, M.; Milani, P.; Bernuzzi, S.; Corato, M.; Ceroni, M.; Cashman, J.R.; Cereda, C. Regulation of FMO and PON Detoxication Systems in ALS Human Tissues. Neurotox. Res. 2012, 23, 370–377. [Google Scholar] [CrossRef]
- Rosenblat, M.; Volkova, N.; Roqueta-Rivera, M.; Nakamura, M.T.; Aviram, M. Increased macrophage cholesterol biosynthesis and decreased cellular paraoxonase 2 (PON2) expression in Δ6-desaturase knockout (6-DS KO) mice: Beneficial effects of arachidonic acid. Atherosclerosis 2010, 210, 414–421. [Google Scholar] [CrossRef]
- Rosenblat, M.; Hayek, T.; Hussein, K.; Aviram, M. Decreased Macrophage Paraoxonase 2 Expression in Patients with Hypercholesterolemia Is the Result of Their Increased Cellular Cholesterol Content: Effect of Atorvastatin Therapy. Arter. Thromb. Vasc. Biol. 2004, 24, 175–180. [Google Scholar] [CrossRef] [Green Version]
- Lim, J.A.; Kim, S.H. Transcriptional activation of an anti-oxidant mouse Pon2 gene by dexamethasone. BMB Rep. 2009, 42, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Boesch-Saadatmandi, C.; Pospissil, R.T.; Graeser, A.-C.; Canali, R.; Boomgaarden, I.; Doering, F.; Wolffram, S.; Egert, S.; Mueller, M.J.; Rimbach, G. Effect of Quercetin on Paraoxonase 2 Levels in RAW264.7 Macrophages and in Human Monocytes—Role of Quercetin Metabolism. Int. J. Mol. Sci. 2009, 10, 4168–4177. [Google Scholar] [CrossRef]
- Fernandes, E.S.; Machado, M.D.O.; Becker, A.M.; De Andrade, F.; Maraschin, M.; Da Silva, E.L. Yerba mate (Ilex paraguariensis) enhances the gene modulation and activity of paraoxonase-2: In vitro and in vivo studies. Nutrients 2012, 28, 1157–1164. [Google Scholar] [CrossRef] [PubMed]
- Ravi, R.; Nagaraj, N.R.; Rajesh, B.S.; Ramya, R.; Nareshkumar, R.N.; Bharathidevi, S. Effect of advanced glycation end product on paraoxonase 2 expression: Its impact on endoplasmic reticulum stress and inflammation in HUVECs. Life Sci. 2020, 246, 117397. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Esparragón, F.; Fernández, J.C.L.; Ríos, N.B.; García-Bello, M.A.; Velazquez, E.H.; Cappiello, L.; Pérez, J.C.R. Paraoxonase 1 and 2 gene variants and the ischemic stroke risk in Gran Canaria population: An association study and meta-analysis. Int. J. Neurosci. 2016, 127, 191–198. [Google Scholar] [CrossRef]
- Wu, S.S.; Yu, J.N.; Jiao, J.; Chen, G.S.; Zhang, C.Y.; Yu, S.F. Association between PON2 gene polymorphisms and susceptibility to noise-induced hearing loss. Chin. J. Ind. Hyg. Occup. Dis. 2020, 38, 128–132. [Google Scholar]
- Li, X.; Cao, J.; Wang, J.; Song, H.; Ji, G.; Dong, Q.; Wei, C.; Cao, Y.; Wang, B.; Zhu, B.; et al. PON2 and ATP2B2 gene polymorphisms with noise-induced hearing loss. J. Thorac. Dis. 2016, 8, 430–438. [Google Scholar] [CrossRef] [Green Version]






| Disease | References | ||
|---|---|---|---|
| Heart Diseases | |||
| PON2 | Atherosclerosis | Devarajan, A., et al., 2011 | [2] |
| Ng, C.J., et al., 2001 | [14] | ||
| Ng, C.J., et al., 2005 | [15] | ||
| Ng, C.J., et al., 2006a | [97] | ||
| Ng, C.J., et al., 2006b | [112] | ||
| Acute myocardial ischemia | Marchegiani, et al., 2009 | [42] | |
| S311CSNP | Sulaiman, D., et al., 2019 | [103] | |
| Coronary Artery Disease (CAD) | Li, W., et al., 2018 | [5] | |
| PON2 SNPs | Wang, X., et al., 2003 Martinelli, N., et al., 2004 | [31] [32] | |
| Sanghera, D.K., et al., 1998 | [33] | ||
| Shin, B.S., et al., 2008 | [38] | ||
| Chen, Q., et al., 2003 | [39] | ||
| Chen, M., et al., 2016 | [41] | ||
| Leus, F.R., et al., 2001 | [45] | ||
| Large vessel disease | Slowik, A., et al., 2007 | [43] | |
| PON2SNP | Covid-19 (blood coagulation) | Liu, D., et al., 2020 | [125] |
| Metabolic Diseases | |||
| PON2 | Insulin sensitivity | Qujeq, D., et al., 2018 | [50] |
| Bourquard, N., et al., 2011 | [107] | ||
| PON2 PON2SNPs | Diabetes complications | Pinizzotto, M., et al., 2001 | [49] |
| Mackness, B., et al., 2005 | [51] | ||
| Cancer | |||
| PON2 | Different types of malignancies | Witte, I., et al., 2011 | [4] |
| Shakhparonov, M.I., et al., 2018 | [6] | ||
| Witte, I., et al., 2012 | [141] | ||
| Bacchetti, T., et al., 2019 | [142] | ||
| Hepatocellular carcinoma | Yao, L., et al., 2002 | [143] | |
| Prostate carcinoma | Ribarska, T., et al., 2010 | [144] | |
| Skin neoplasms | Bacchetti, T., et al., 2020 | [145] | |
| Gastric cancer | Wang, X., et al., 2019 | [146] | |
| Breast cancer | Wang, R., et al., 2018 | [147] | |
| Pediatric acute lymphoblastic leukemia (ALL) | Ross, M.E., et al., 2003 Kang, H., et al., 2010 | [148] [149] | |
| Chronic myeloid leukemia | Frank, O., et al., 2006 | [150] | |
| Human T-cell leukemia virus 1 | Pise-Masison, C.A., et al., 2002 | [151] | |
| Pancreatic cancer | Nagarajan, A., et al., 2017 | [56] | |
| Bladder cancer | Bacchetti, T., et al., 2017 | [152] | |
| Glioblastoma | Tseng, J.H., et al., 2017 | [153] | |
| Oral squamous cell carcinoma (OSCC) | Krüger, M., et al., 2016 Krüger, M., et al., 2015 | [60] [157] | |
| Ovarian tumor | Devarajan, A., et al., 2018 | [158] | |
| Neurodegenerative diseases | |||
| PON2 | Neuroprotection | Costa, L.G., et al., 2014 | [102] |
| Alzheimer | Shi, J.; et al., 2004 | [35] | |
| Erlich, P.M., et al., 2006 | [46] | ||
| Nie, Y., et al., 2017 | [167] | ||
| Leduc, V., et al., 2011 | [168] | ||
| Mu, N., et al., 2013 | [169] | ||
| Janka, Z., et al., 2002 | [34] | ||
| PON2 SNPs | Parkinson | Parsanejad, M., et al., 2014 | [180] |
| Aguirre-Vidal, Y., et al., 2020 | [183] | ||
| ALS | Narain, P., et al., 2019 | [185] | |
| Saeed, M., et al., 2006 | [188] | ||
| Landers, J.E., et al., 2008 | [190] | ||
| Valdmanis, P.N., et al., 2008 | [191] | ||
| Ticozzi, N., et al., 2010 | [192] | ||
| Ricci, C., et al., 2011 | [193] | ||
| Chen, Y.P., et al., 2012 | [194] | ||
| Other diseases | |||
| PON2 SNPs | Cataract | Baig, A., et al., 2019 | [40] |
| PON2 | Ischemic stroke | Rodríguez-E, F., et al., 2017 | [202] |
| PON2 SNPs | Noise-induced hearing loss | Wu, S.S., et al., 2020 | [203] |
| Li, X., et al., 2016 | [204] | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manco, G.; Porzio, E.; Carusone, T.M. Human Paraoxonase-2 (PON2): Protein Functions and Modulation. Antioxidants 2021, 10, 256. https://doi.org/10.3390/antiox10020256
Manco G, Porzio E, Carusone TM. Human Paraoxonase-2 (PON2): Protein Functions and Modulation. Antioxidants. 2021; 10(2):256. https://doi.org/10.3390/antiox10020256
Chicago/Turabian StyleManco, Giuseppe, Elena Porzio, and Teresa Maria Carusone. 2021. "Human Paraoxonase-2 (PON2): Protein Functions and Modulation" Antioxidants 10, no. 2: 256. https://doi.org/10.3390/antiox10020256
APA StyleManco, G., Porzio, E., & Carusone, T. M. (2021). Human Paraoxonase-2 (PON2): Protein Functions and Modulation. Antioxidants, 10(2), 256. https://doi.org/10.3390/antiox10020256