Tracing the Evolution of Plant Glyoxalase III Enzymes for Structural and Functional Divergence
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Retrieval and Sequence Analysis
2.2. Phylogenetic and Subcellular Localization Analysis
2.3. Motif and Three-Dimensional Structural Analysis of GLYIII Proteins
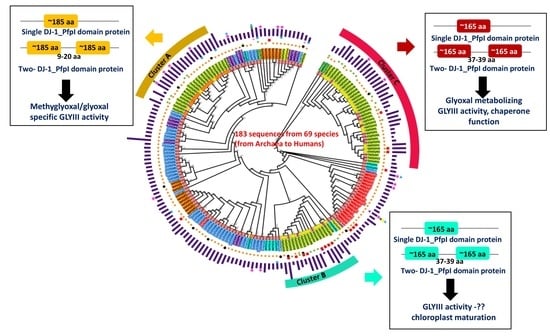
3. Results
3.1. GLYIII Proteins Contain DJ-1_Pfp1 Domain in Combination with Various Additional Domains
3.2. Plants Have Three Major Types of GLYIII Proteins
3.3. Cluster-C GLYIII Proteins Share Similarity with Their Ancestral Homologs in Localization Patterns
3.4. Assessment of Domain Relationships of GLYIII Proteins Indicate Primitive Evolution of N-Terminal Domain of Plant GLYIII
3.5. Extensive Motif Rearrangements Have Shaped the Evolution of Plant GLYIII Proteins
3.6. Assessment of Motif Organization in DJ-1_PfpI Domains Reveals Both Vertical and Horizontal Evolutionary Patterns of Plant GLYIII Domains
3.7. Variations in the Three-Dimensional Predicted Structure of Plant GLYIII Proteins from Different Clusters Are Indicative of Their Functional Flexibility
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GLY | glyoxalase |
| GSH | glutathione |
| MG | methylglyoxal |
References
- Kalapos, M.P.; Biology, T. Methylglyoxal in living organisms—Chemistry, biochemistry, toxicology and biological implications. Toxicol. Lett. 1999, 110, 145–175. [Google Scholar] [CrossRef]
- Kalapos, M.P. The tandem of free radicals and methylglyoxal. Chem. Biol. Interact. 2008, 171, 251–271. [Google Scholar] [CrossRef]
- Thornalley, P.J. Pharmacology of methyglyoxal: Formation, modification of proteins and nucleic acids, and enzymatic detoxification—A role in pathogenesis and antiproliferative chemotherapy. Gen. Pharmacol. 1996, 27, 565–573. [Google Scholar] [CrossRef]
- Thornalley, P.J. Protein and nucleotide damage by glyoxal and methylglyoxal in physiological systems—role in ageing and disease. Drug Metabol. Drug Interact. 2008, 23, 125–150. [Google Scholar] [CrossRef] [PubMed]
- Singla-Pareek, S.L.; Reddy, M.K.; Sopory, S.K. Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proc. Natl. Acad. Sci. USA 2003, 100, 14672–14677. [Google Scholar] [CrossRef]
- Kaur, C.; Vishnoi, A.; Ariyadasa, T.U.; Bhattacharya, A.; Singla-Pareek, S.L.; Sopory, S.K. Episodes of horizontal gene-transfer and gene-fusion led to co-existence of different metal-ion specific glyoxalase I. Sci. Rep. 2013, 3, 3076. [Google Scholar] [CrossRef]
- Misra, K.; Banerjee, A.B.; Ray, S.; Ray, M. Glyoxalase III from Escherichia coli: A single novel enzyme for the conversion of methylglyoxal into D-lactate without reduced glutathione. Biochem. J. 1995, 305, 999–1003. [Google Scholar] [CrossRef]
- Zhao, Q.; Su, Y.; Wang, Z.; Chen, C.; Wu, T.; Huang, Y. Identification of glutathione (GSH)-independent glyoxalase III from Schizosaccharomyces pombe. BMC Evol. Biol. 2014, 14, 86. [Google Scholar] [CrossRef]
- Ghosh, A.; Kushwaha, H.R.; Hasan, M.R.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. presence of unique glyoxalase iii proteins in plants indicates the existence of shorter route for methylglyoxal detoxification. Sci. Rep. 2016, 6, 18358. [Google Scholar] [CrossRef]
- Mannervik, B.; Ridderstrom, M. Catalytic and molecular properties of glyoxalase I. Biochem. Soc. Trans 1993, 21, 515–517. [Google Scholar] [CrossRef]
- Inoue, Y.; Maeta, K.; Nomura, W. Glyoxalase system in yeasts: Structure, function, and physiology. Semin. Cell Dev. Biol. 2011, 22, 278–284. [Google Scholar] [CrossRef]
- Suttisansanee, U.; Honek, J.F. Bacterial glyoxalase enzymes. Semin. Cell Dev. Biol. 2011, 22, 285–292. [Google Scholar] [CrossRef]
- Martins, A.M.; Cordeiro, C.; Freire, A.P. Glyoxalase II in Saccharomyces cerevisiae: In situ kinetics using the 5,5′-dithiobis(2-nitrobenzoic acid) assay. Arch. Biochem. Biophys. 1999, 366, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, L.; Chin, L.S. Parkinson disease protein DJ-1 converts from a zymogen to a protease by carboxyl-terminal cleavage. Hum. Mol. Genet. 2010, 19, 2395–2408. [Google Scholar] [CrossRef]
- Lewandowska, A.; Vo, T.N.; Nguyen, T.-D.H.D.H.; Wahni, K.; Vertommen, D.; Van Breusegem, F.; Young, D.; Messens, J. Bifunctional chloroplastic DJ-1B from Arabidopsis thaliana is an oxidation-robust holdase and a glyoxalase sensitive to H2O2. Antioxidants 2019, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Richarme, G.; Mihoub, M.; Dairou, J.; Chi Bui, L.; Leger, T.; Lamouri, A. Parkinsonism-associated protein DJ-1/park7 is a major protein deglycase that repairs methylglyoxal- and glyoxal-glycated cysteine, arginine, and lysine residues. J. Biol. Chem. 2015, 290, 1885–1897. [Google Scholar] [CrossRef]
- Richarme, G.; Liu, C.; Mihoub, M.; Abdallah, J.; Leger, T.; Joly, N.; Liebart, J.C.; Jurkunas, U.V.; Nadal, M.; Bouloc, P.; et al. Guanine glycation repair by DJ-1/Park7 and its bacterial homologs. Science 2017, 357, 208–211. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Mayorga, E.; Díaz-Sánchez, Á.G.; Dagda, R.K.R.Y.; Domínguez-Solís, C.A.; Dagda, R.K.R.Y.; Coronado-Ramírez, C.K.; Martínez-Martínez, A. Novel Redox-dependent Esterase activity (EC 3.1.1.2) for DJ-1: Implications for Parkinson’s disease. Int. J. Mol. Sci. 2016, 17, 1346. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.E.; Mouradian, M.M. Cytoprotective mechanisms of DJ-1 against oxidative stress through modulating ERK1/2 and ASK1 signal transduction. Redox Biol. 2018, 14, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Ariga, H.; Takahashi-Niki, K.; Kato, I.; Maita, H.; Niki, T.; Iguchi-Ariga, S.M.M.M. Neuroprotective function of DJ-1 in Parkinson’s disease. Oxid. Med. Cell. Longev. 2013, 2013, 683920. [Google Scholar] [CrossRef] [PubMed]
- Bonifati, V.; Rizzu, P.; Van Baren, M.J.; Schaap, O.; Breedveld, G.J.; Krieger, E.; Dekker, M.C.J.; Squitieri, F.; Ibanez, P.; Joosse, M.; et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science 2003, 299, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.M.; Lin, H.; Maple, J.; Björkblom, B.; Alves, G.; Larsen, J.P.; Møller, S.G. The Arabidopsis DJ-1a protein confers stress protection through cytosolic SOD activation. J. Cell Sci. 2010, 123, 1644–1651. [Google Scholar] [CrossRef]
- Lin, J.; Nazarenus, T.J.; Frey, J.L.; Liang, X.; Wilson, M.A.; Stone, J.M. A plant DJ-1 homolog is essential for Arabidopsis thaliana chloroplast development. PLoS ONE 2011, 6, e23731. [Google Scholar] [CrossRef]
- Melvin, P.; Bankapalli, K.; D’Silva, P.; Shivaprasad, P.V. Methylglyoxal detoxification by a DJ-1 family protein provides dual abiotic and biotic stress tolerance in transgenic plants. Plant Mol. Biol. 2017, 94, 381–397. [Google Scholar] [CrossRef] [PubMed]
- Mohanan, M.V.; Pushpanathan, A.; Sasikumar, S.P.T.; Selvarajan, D.; Jayanarayanan, A.N.; Kumar, A.; Ramalingam, S.; Karuppasamy, S.N.; Subbiah, R.; Ram, B.; et al. Ectopic expression of DJ-1/PfpI domain containing Erianthus arundinaceus Glyoxalase III (EaGly III) enhances drought tolerance in sugarcane. Plant Cell Rep. 2020, 39, 1581–1594. [Google Scholar] [CrossRef] [PubMed]
- Hasim, S.; Hussin, N.A.; Alomar, F.; Bidasee, K.R.; Nickerson, K.W.; Wilson, M.A. A glutathione-independent glyoxalase of the DJ-1 superfamily plays an important role in managing metabolically generated methylglyoxal in Candida albicans. J. Biol. Chem. 2014, 289, 1662–1674. [Google Scholar] [CrossRef] [PubMed]
- Singla-Pareek, S.L.; Kaur, C.; Kumar, B.; Pareek, A.; Sopory, S.K. Reassessing plant glyoxalases: Large family and expanding functions. New Phytol. 2020, 227, 714–721. [Google Scholar] [CrossRef]
- Kaur, C.; Sharma, S.; Hasan, M.R.; Pareek, A.; Singla-Pareek, S.L.; Sopory, S.K. Characteristic variations and similarities in biochemical, molecular, and functional properties of glyoxalases across prokaryotes and eukaryotes. Int. J. Mol. Sci. 2017, 18, 250. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 5 May 2020).
- Phytozome v12.1: Home. Available online: https://phytozome.jgi.doe.gov/pz/portal.html (accessed on 5 May 2020).
- Letunic, I.; Doerks, T.; Bork, P. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015, 43, D257–D260. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.S.; Lin, C.J.; Hwang, J.K. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n -peptide compositions. Protein Sci. 2004, 13, 1402–1406. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.S.; Chen, Y.C.; Lu, C.H.; Hwang, J.K. Prediction of protein subcellular localization. Proteins Struct. Funct. Genet. 2006, 64, 643–651. [Google Scholar] [CrossRef]
- Bannai, H.; Tamada, Y.; Maruyama, O.; Nakai, K.; Miyano, S. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 2002, 18, 298–305. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef]
- DeLano, W.L. The PyMOL Molecular Graphics System. 2002. Available online: http://www.pymol.org (accessed on 27 March 2020).
- Bai, J.; Cederbaum, A.I. Mitochondrial catalase and oxidative injury. Neurosignals 2001, 10, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Guy, B.; Krell, T.; Sanchez, V.; Kennel, A.; Manin, C.; Sodoyer, R. Do Th1 or Th2 sequence motifs exist in proteins? Identification of amphipatic immunomodulatory domains in Helicobacter pylori catalase. Immunol. Lett. 2005, 96, 261–275. [Google Scholar] [CrossRef]
- Kwon, K.; Choi, D.; Hyun, J.K.; Jung, H.S.; Baek, K.; Park, C. Novel glyoxalases from Arabidopsis thaliana. FEBS J. 2013, 280, 3328–3339. [Google Scholar] [CrossRef] [PubMed]
- Subedi, K.P.; Choi, D.; Kim, I.; Min, B.; Park, C. Hsp31 of Escherichia coli K-12 is glyoxalase III. Mol. Microbiol. 2011, 81, 926–936. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.A. The role of cysteine oxidation in DJ-1 function and dysfunction. Antioxidants Redox Signal. 2011, 15, 111–122. [Google Scholar] [CrossRef]
- Van Der Brug, M.P.; Blackinton, J.; Chandran, J.; Hao, L.Y.; Lal, A.; Mazan-Mamczarz, K.; Martindale, J.; Xie, C.; Ahmad, R.; Thomas, K.J.; et al. RNA binding activity of the recessive parkinsonism protein DJ-1 supports involvement in multiple cellular pathways. Proc. Natl. Acad. Sci. USA 2008, 105, 10244–10249. [Google Scholar] [CrossRef]
- Lee, C.; Lee, J.; Lee, J.; Park, C. Characterization of the Escherichia coli YajL, YhbO and ElbB glyoxalases. FEMS Microbiol. Lett. 2016, 363, fnv239. [Google Scholar] [CrossRef]
- Bankapalli, K.; Saladi, S.D.; Awadia, S.S.; Goswami, A.V.; Samaddar, M.; D’Silva, P. Robust Glyoxalase activity of Hsp31, a ThiJ/DJ-1/PfpI Family Member Protein, Is Critical for Oxidative Stress Resistance in Saccharomyces cerevisiae. J. Biol. Chem. 2015, 290, 26491–26507. [Google Scholar] [CrossRef]
- Junn, E.; Jang, W.H.; Zhao, X.; Jeong, B.S.; Mouradian, M.M. Mitochondrial localization of DJ-1 leads to enhanced neuroprotection. J. Neurosci. Res. 2009, 87, 123–129. [Google Scholar] [CrossRef]
- Lei, Y.; Zhang, Z.F.; Lei, R.X.; Wang, S.; Zhuang, Y.; Liu, A.C.; Wu, Y.; Chen, J.; Tang, J.C.; Pan, M.X.; et al. DJ-1 Suppresses Cytoplasmic TDP-43 Aggregation in Oxidative Stress-Induced Cell Injury. J. Alzheimer’s Dis. 2018, 66, 1001–1014. [Google Scholar] [CrossRef]
- Im, J.Y.; Lee, K.W.; Woo, J.-M.; Junn, E.; Mouradian, M.M. DJ-1 induces thioredoxin 1 expression through the Nrf2 pathway. Hum. Mol. Genet. 2012, 21, 3013–3024. [Google Scholar] [CrossRef] [PubMed]
- Canet-Avilés, R.M.; Wilson, M.A.; Miller, D.W.; Ahmad, R.; McLendon, C.; Bandyopadhyay, S.; Baptista, M.J.; Ringe, D.; Petsko, G.A.; Cookson, M.R. The Parkinson’s disease DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc. Natl. Acad. Sci. USA 2004, 101, 9103–9108. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Shimoji, M.; Thomas, B.; Moore, D.J.; Yu, S.-W.; Marupudi, N.I.; Torp, R.; Torgner, I.A.; Ottersen, O.P.; Dawson, T.M.; et al. Mitochondrial localization of the Parkinson’s disease related protein DJ-1: Implications for pathogenesis. Hum. Mol. Genet. 2005, 14, 2063–2073. [Google Scholar] [CrossRef]
- Bankapalli, K.; Vishwanathan, V.; Susarla, G.; Sunayana, N.; Saladi, S.D.; Peethambaram, D.; D’Silva, P. Redox-dependent regulation of mitochondrial dynamics by DJ-1 paralogs in Saccharomyces cerevisiae: Regulation of mitochondrial health by DJ-1 homologs. Redox Biol. 2020, 32, 101451. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, B.; Kaur, C.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. Tracing the Evolution of Plant Glyoxalase III Enzymes for Structural and Functional Divergence. Antioxidants 2021, 10, 648. https://doi.org/10.3390/antiox10050648
Kumar B, Kaur C, Pareek A, Sopory SK, Singla-Pareek SL. Tracing the Evolution of Plant Glyoxalase III Enzymes for Structural and Functional Divergence. Antioxidants. 2021; 10(5):648. https://doi.org/10.3390/antiox10050648
Chicago/Turabian StyleKumar, Brijesh, Charanpreet Kaur, Ashwani Pareek, Sudhir K. Sopory, and Sneh L. Singla-Pareek. 2021. "Tracing the Evolution of Plant Glyoxalase III Enzymes for Structural and Functional Divergence" Antioxidants 10, no. 5: 648. https://doi.org/10.3390/antiox10050648
APA StyleKumar, B., Kaur, C., Pareek, A., Sopory, S. K., & Singla-Pareek, S. L. (2021). Tracing the Evolution of Plant Glyoxalase III Enzymes for Structural and Functional Divergence. Antioxidants, 10(5), 648. https://doi.org/10.3390/antiox10050648