Nanomechanical Study of Enzyme: Coenzyme Complexes: Bipartite Sites in Plastidic Ferredoxin-NADP+ Reductase for the Interaction with NADP+
Abstract
:1. Introduction
2. Materials and Methods
2.1. Protein Labeling and Immobilization of FNR on Mica
2.2. Functionality of the Enzyme Samples
2.3. AFM Tip Functionalization with NADP+
2.4. Dynamic Force Spectroscopy Measurements
2.5. Analysis of the Force Curves: Mechanostability Parameters
3. Results
3.1. Analysis of the Enzymatic Samples
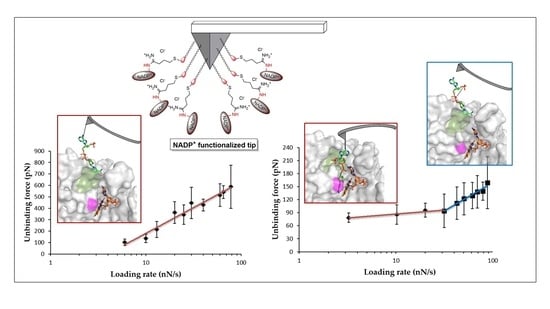
3.2. Mechanical Stability of the FNR Complexes
3.3. Dissociation Kinetics for AnFNRox:NADP+ Complexes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zinn, M.; Egli, T. Occurrence Synthesis and Medical Application of Bacterial Polyhydroxyalkanoate. Adv. Drug Deliv. Rev. 2001, 53, 5–21. [Google Scholar] [CrossRef]
- Lin, H.; Gao, W.; Meng, F.; Liao, B.; Leung, K.; Zhao, L.; Chen, J.; Hong, H. Membrane Bioreactors for Industrial Wastewater Treatment: A Critical Review. Crit. Rev. Environ. Sci. Technol. 2012, 42, 677–740. [Google Scholar] [CrossRef]
- Ando, T. High-speed atomic force microscopy and its future prospects. Biophys. Rev. 2018, 10, 285–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lira-Navarrete, E.; Pallares, M.C.; Castello, F.; Ruedas-Rama, M.J.; Orte, A.; Lostao, A.; Hurtado-Guerrero, R. Protein O-Fucosyltransferase 1 Undergoes Interdomain Flexibility in Solution. Molecules 2021, 26, 2105. [Google Scholar] [CrossRef]
- Marcuello, C.; Arilla-Luna, S.; Medina, M.; Lostao, A. Detection of a quaternary organization into dimer of trimers of Corynebacterium ammoniagenes FAD synthetase at the single-molecule level and at the in cell level. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2013, 1834, 665–676. [Google Scholar] [CrossRef]
- Pallarés, M.C.; Marcuello, C.; Botello-Morte, L.; González, A.; Fillat, M.F.; Lostao, A. Sequential binding of FurA from Anabaena sp. PCC 7120 to iron boxes: Exploring regulation at the nanoscale. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2014, 1844, 623–631. [Google Scholar] [CrossRef]
- Lira-Navarrete, E.; de las Rivas, M.; Compañón, I.; Pallarés, M.C.; Kong, Y.; Iglesias-Fernández, J.; Bernardes, G.J.L.; Peregrina, J.M.; Rovira, C.; Bernardó, P.; et al. Dynamic interplay between catalytic and lectin domains of GalNAc-transferases modulates protein O-glycosylation. Nat. Commun. 2015, 6, 6937. [Google Scholar] [CrossRef] [Green Version]
- Neuman, K.C.; Nagy, A. Single-molecule force spectroscopy: Optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods 2008, 5, 491–505. [Google Scholar] [CrossRef]
- Binnig, G.; Quate, C.F.; Gerber, C. Atomic Force Microscope. Phys. Rev. Lett. 1986, 56, 930–933. [Google Scholar] [CrossRef] [Green Version]
- Florin, E.L.; Moy, V.T.; Gaub, H.E. Adhesion forces between individual ligand-receptor pairs. Science 1994, 264, 415–417. [Google Scholar] [CrossRef] [Green Version]
- Merkel, R.; Nassoy, P.; Leung, L.; Ritchie, K.; Evans, E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature 1999, 397, 50–53. [Google Scholar] [CrossRef] [PubMed]
- Pierres, A.; Touchard, D.; Benoliel, A.M.; Bongrand, P. Dissecting streptavidin-biotin interaction with a laminar flow chamber. Biophys. J. 2002, 82, 3214–3223. [Google Scholar] [CrossRef] [Green Version]
- Capitanio, M.; Pavone, F.S. Interrogating biology with force: Single molecule high-resolution measurements with Optical Tweezers. Biophys. J. 2013, 105, 1293–1303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarkar, R.; Rybenkov, V.V. A guide to Magnetic Tweezers and their applications. Front. Phys. 2016, 4, 48. [Google Scholar] [CrossRef]
- Williams, M.C.; Rouzina, I. Force spectroscopy of single DNA and RNA molecules. Curr. Opin. Struct. Biol. 2002, 12, 330–336. [Google Scholar] [CrossRef]
- Puchner, E.M.; Gaub, H.E. Force and function: Probing proteins with AFM-based force spectroscopy. Curr. Opin. Struct. Biol. 2009, 19, 605–614. [Google Scholar] [CrossRef]
- Alegre-Cebollada, J.; Perez-Jimenez, R.; Kosuri, P.; Fernandez, J.M. Single-molecule Force Spectroscopy approach to enzyme catalysis. J. Biol. Chem. 2010, 285, 18961–18966. [Google Scholar] [CrossRef] [Green Version]
- Medina, M.; Gomez-Moreno, C. Interaction of ferredoxin-NADP(+) reductase with its substrates: Optimal interaction for efficient electron transfer. Photosynth. Res. 2004, 79, 113–131. [Google Scholar] [CrossRef]
- Medina, M. Structural and mechanistic aspects of flavoproteins: Photosynthetic electron transfer from photosystem I to NADP+. FEBS J. 2009, 276, 3942–3958. [Google Scholar] [CrossRef]
- Mulo, P.; Medina, M. Interaction and electron transfer between ferredoxin-NADP+ oxidoreductase and its partners: Structural, functional, and physiological implications. Photosynth. Res. 2017, 134, 265–280. [Google Scholar] [CrossRef]
- Hermoso, J.A.; Mayoral, T.; Faro, M.; Gomez-Moreno, C.; Sanz-Aparicio, J.; Medina, M. Mechanism of coenzyme recognition and binding revealed by crystal structure analysis of ferredoxin-NADP+ reductase complexed with NADP+. J. Mol. Biol. 2002, 319, 1133–1142. [Google Scholar] [CrossRef]
- Medina, M.; Luquita, A.; Tejero, J.; Hermoso, J.; Mayoral, T.; Sanz-Aparicio, J.; Grever, K.; Gómez-Moreno, C. Probing the determinants of coenzyme specificity in ferredoxin-NADP+ reductase by site-directed mutagenesis. J. Biol. Chem. 2001, 276, 11902–11912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tejero, J.; Martínez-Júlvez, M.; Mayoral, T.; Luquita, A.; Sanz-Aparicio, J.; Hermoso, J.A.; Hurley, J.K.; Tollin, G.; Gómez-Moreno, C.; Medina, M. Involvement of the pyrophosphate and the 2′-phosphate binding regions of ferredoxin-NADP+ reductase in coenzyme specificity. J. Biol. Chem. 2003, 278, 49203–49214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, Z.; Aliverti, A.; Zanetti, G.; Arakaki, A.K.; Ottado, J.; Orellano, E.G.; Calcaterra, N.B.; Ceccarelli, E.A.N.; Carrillo, N.; Karplus, P.A. A productive NADP+ binding mode of ferredoxin-NADP+ reductase revealed by protein engineering and crystallographic studies. Nat. Struct. Biol. 1999, 6, 847–853. [Google Scholar] [CrossRef] [PubMed]
- Carrillo, N.; Ceccarelli, E.A. Open questions in ferredoxin-NADP+ reductase catalytic mechanism. Eur. J. Biochem. 2003, 270, 1900–1915. [Google Scholar] [CrossRef] [PubMed]
- Peregrina, J.R.; Sánchez-Azqueta, A.; Herguedas, B.; Martínez-Júlvez, M.; Medina, M. Role of specific residues in coenzyme binding, charge-transfer complex formation, and catalysis in Anabaena ferredoxin-NADP+ reductase. Biochim. Biophys. Acta (BBA)-Bioenerg. 2010, 1797, 1638–1646. [Google Scholar] [CrossRef] [Green Version]
- Peregrina, J.R.; Lans, I.; Medina, M. The transient catalytically competent coenzyme allocation into the active site of Anabaena ferredoxin NADP+ -reductase. Eur. Biophys. J. 2012, 41, 117–128. [Google Scholar] [CrossRef]
- Piubelli, L.; Aliverti, A.; Arakaki, A.K.; Carrillo, N.; Ceccarelli, E.A.; Karplus, P.A.; Zanetti, G. Competition between C-terminal tyrosine and nicotinamide modulates pyridine nucleotide affinity and specificity in plant ferredoxin-NADP+ reductase. J. Biol. Chem. 2000, 275, 10472–10476. [Google Scholar] [CrossRef] [Green Version]
- Tejero, J.; Pérez-Dorado, I.; Maya, C.; Martínez-Júlvez, M.; Sanz-Aparicio, J.; Gómez-Moreno, C.; Hermoso, J.A.; Medina, M. C-Terminal Tyrosine of Ferredoxin−NADP+ Reductase in Hydride Transfer Processes with NAD(P)+/H. Biochemistry 2005, 44, 13477–13490. [Google Scholar] [CrossRef]
- Lans, I.; Medina, M.; Rosta, E.; Hummer, G.; Garcia-Viloca, M.; Lluch, J.M.; González-Lafont, À. Theoretical study of the mechanism of the hydride transfer between ferredoxin-NADP+ reductase and NADP+: The role of Tyr303. J. Am. Chem. Soc. 2012, 134, 20544–20553. [Google Scholar] [CrossRef]
- Marcuello, C.; Frempong, G.A.; Balsera, M.; Medina, M.; Lostao, A. Atomic Force Microscopy to Elicit Conformational Transitions of Ferredoxin-Dependent Flavin Thioredoxin Reductases. Antioxidants 2021, 10, 1437. [Google Scholar] [CrossRef] [PubMed]
- Marcuello, C.; Miguel, R.; Medina, M.; Martínez-Júlvez, M.; Gómez-Moreno, C.; Lostao, A. Nanomechanics of ferredoxin-NADP(+) reductase complexes analyzed by AFM single-molecule force spectroscopy. FEBS J. 2012, 279, 513. [Google Scholar]
- Marcuello, C.; de Miguel, R.; Martínez-Júlvez, M.; Gómez-Moreno, C.; Lostao, A. Mechanostability of the Single-Electron-Transfer Complexes of Anabaena Ferredoxin-NADP(+) Reductase. ChemPhysChem 2015, 16, 3161–3169. [Google Scholar] [CrossRef] [PubMed]
- Marcuello, C.; de Miguel, R.; Martínez-Júlvez, M.; Gómez-Moreno, C.; Lostao, A. Inside Cover: Mechanostability of the single electron-transfer complexes of Anabaena ferredoxin–NADP+ reductase. ChemPhysChem 2015, 16, 3120. [Google Scholar] [CrossRef] [Green Version]
- Marcuello, C.; de Miguel, R.; Gómez-Moreno, C.; Martínez-Júlvez, M.; Lostao, A. An efficient method for enzyme immobilization evidenced by atomic force microscopy. Protein Eng. Des. Sel. 2012, 25, 715–723. [Google Scholar] [CrossRef] [Green Version]
- Gómez-Moreno, C.; Lostao, A. Single molecule methods to study flavoproteins. In Flavoproteins, Dehydrogenases and Physical Methods; Hille, R., Miller, S., Palfey, B., Eds.; De Gruyter: Berlin, Germany, 2013; Volume 2, pp. 297–318. [Google Scholar] [CrossRef]
- Gómez-Moreno, C.; Marcuello, C.; de Miguel, R.; Medina, M.; Martínez-Júlvez, M.; Lostao, A. Atomic Force Microscopy as tool to understand the Mechanoestability of Ferredoxin-NADP+ reductase Complexes with its partners. In Flavins and Flavoproteins; Miller, S., Hille, R., Palfey, B., Eds.; Lulu.com.: Raleigh, NC, USA, 2011; Volume 17, pp. 569–575. [Google Scholar]
- Horcas, I.; Fernandez, R.; Gomez-Rodriguez, J.M.; Colchero, J.; Gomez-Herrero, J.; Baro, A.M. WSXM: A software for scanning probe microscopy and a tool for nanotechnology. Rev. Sci. Instrum. 2007, 78, 13705. [Google Scholar] [CrossRef]
- Lostao, A.; Medina, M. Atomic Force Microscopy: Single-Molecule Imaging and Force Spectroscopy in the Study of Flavoproteins Ligand Binding and Reaction Mechanisms. Methods Mol. Biol. 2021, 2280, 157–178. [Google Scholar] [CrossRef]
- Bouchiat, C.; Wang, M.D.; Allemand, J.; Strick, T.; Blocks, S.M.; Croquette, V. Estimating the persistence length of a worm-like chain molecule from force-extension measurements. Biophys. J. 1999, 76, 409–413. [Google Scholar] [CrossRef] [Green Version]
- Tapia-Rojo, R.; Marcuello, C.; Lostao, A.; Gómez-Moreno, C.; Mazo, J.; Falo, F. A physical picture for mechanical dissociation of biological complexes: From forces to free energies. Phys. Chem. Chem. Phys. 2017, 19, 4567–4575. [Google Scholar] [CrossRef] [Green Version]
- Evans, E.; Ritchie, K. Dynamic strength of molecular adhesion bonds. Biophys. J. 1997, 72, 1541–1555. [Google Scholar] [CrossRef] [Green Version]
- Evans, E. Probing the relation between force—lifetime—and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 2001, 30, 105–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, C.; Chen, A.; Kolb, P.; Moy, V.T. Energy landscape of streptavidin-biotin complexes measured by atomic force microscopy. Biochemistry 2000, 39, 10219–10223. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, G.; Haran, G.; Zhou, H.-X. Fundamental aspects of protein–protein association kinetics. Chem. Rev. 2009, 109, 839–860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonanni, B.; Bizzarri, A.R.; Cannistraro, S. Optimized Biorecognition of Cytochrome c551 and Azurin Immobilized on Thiol-Terminated Monolayers Assembled on Au (111) Substrates. J. Phys. Chem. 2006, 110, 14574–14580. [Google Scholar] [CrossRef]
- Funari, G.; Domenici, F.; Nardinocchi, L.; Puca, R.; D’Orazi, G.; Bizarri, A.R.; Cannistraro, S. Interaction of p53 with Mdm2 and azurin as studied by atomic force spectroscopy. J. Mol. Recognit. 2010, 23, 343–351. [Google Scholar] [CrossRef]
- Fritz, J.; Katopodis, A.G.; Kolbinger, F.; Anselmetti, D. Force-mediated kinetics of single P-selectin ligand complexes observed by atomic force microscopy. Proc. Natl. Acad. Sci. USA 1998, 95, 12283–12288. [Google Scholar] [CrossRef] [Green Version]
- Neuert, G.; Albrecht, C.; Pamir, E.; Gaub, H.E. Dynamic force spectroscopy of the digoxigenin-antibody complex. FEBS Lett. 2006, 580, 505–509. [Google Scholar] [CrossRef] [Green Version]
- Taranta, M.; Bizzarri, A.R.; Cannistraro, S. Probing the interaction between p53 and the bacterial protein azurin by single molecule force spectroscopy. J. Mol. Recognit. 2008, 21, 63–70. [Google Scholar] [CrossRef]
- Carvalho, F.A.; Connell, S.; Miltenberger-Miltenyi, G.; Pereira, S.V.; Tavares, A.; Ariens, R.A.S.; Santos, N.C. Atomic force microscope-based molecular recognition of a fibrinogen receptor on human erythrocytes. ACS Nano 2010, 4, 4609–4620. [Google Scholar] [CrossRef]
- Teulon, J.M.; Delcuze, Y.; Odorico, M.; Chen, S.W.; Parot, P.; Pellequer, J.L. Single and multiple bonds in (Strept)avidin–biotin interactions. J. Mol. Recogn. 2011, 24, 490–502. [Google Scholar] [CrossRef]
- Lans, I.; Peregrina, J.R.; Medina, M.; Garcia-Viloca, M.; González-Lafont, A.; Lluch, J.M. Mechanism of the hydride transfer between Anabaena Tyr303Ser FNR(rd)/FNR(ox) and NADP+/H. A combined pre-steady-state kinetic/ensemble-averaged transition-state theory with multidimensional tunneling study. J. Phys. Chem. B 2010, 114, 3368–3379. [Google Scholar] [CrossRef] [PubMed]
- Yersin, A.; Osada, T.; Ikai, A. Exploring transferrin-receptor interactions at the single-molecule level. Biophys. J. 2008, 94, 230–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez-Azqueta, A.; Catalano-Dupuy, D.L.; Lopez-Rivero, A.; Tondo, M.L.; Orellano, E.G.; Ceccarelli, E.A.; Medina, M. Dynamics of the active site architecture in plant-type ferredoxin-NADP(+) reductases catalytic complexes. Biochim. Biophys. Acta (BBA)-Bioenerg. 2014, 1837, 1730–1738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monchietti, P.; López Rivero, A.S.; Ceccarelli, E.A.; Catalano-Dupuy, D.L. A new catalytic mechanism of bacterial ferredoxin-NADP+ reductases due to a particular NADP+ binding mode. Protein Sci. 2021, 30, 2106–2120. [Google Scholar] [CrossRef]












| Complex | Unbinding Force (pN) * | Events | koff (s−1) | τ (s) | xβ (nm) |
|---|---|---|---|---|---|
| WT FNRox:NADP+ | 135.7 ± 36.4 | 1 barrier | 0.02020 | 49.42 | 0.021 |
| Y303S FNRox:NADP+ | 85.7 ± 22.4 | inner barrier outer barrier | 0.10490 | 9.53 | 0.073 |
| 0.00002 | 50,838.84 | 0.524 | |||
| Y303F FNRox:NADP+ | 74.4 ± 17.0 | inner barrier outer barrier | 0.01810 | 56.13 | 0.157 |
| 0.09410 | 10.61 | 0.071 | |||
| Y303W FNRox:NADP+ | 68.0 ± 14.7 | 1 barrier | 0.01020 | 98.29 | 0.267 |
| WT FNRox:Fdox | 57 ± 16 | 1 barrier | 21.2 | 0.047 | 0.27 |
| WT FNRox:Fldox | 21 ± 8 | inner barrier | 253.3 | 0.004 | 0.18 |
| outer barrier | 55.7 | 0.018 | 0.47 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Domínguez, S.; Caballero-Mancebo, S.; Marcuello, C.; Martínez-Júlvez, M.; Medina, M.; Lostao, A. Nanomechanical Study of Enzyme: Coenzyme Complexes: Bipartite Sites in Plastidic Ferredoxin-NADP+ Reductase for the Interaction with NADP+. Antioxidants 2022, 11, 537. https://doi.org/10.3390/antiox11030537
Pérez-Domínguez S, Caballero-Mancebo S, Marcuello C, Martínez-Júlvez M, Medina M, Lostao A. Nanomechanical Study of Enzyme: Coenzyme Complexes: Bipartite Sites in Plastidic Ferredoxin-NADP+ Reductase for the Interaction with NADP+. Antioxidants. 2022; 11(3):537. https://doi.org/10.3390/antiox11030537
Chicago/Turabian StylePérez-Domínguez, Sandra, Silvia Caballero-Mancebo, Carlos Marcuello, Marta Martínez-Júlvez, Milagros Medina, and Anabel Lostao. 2022. "Nanomechanical Study of Enzyme: Coenzyme Complexes: Bipartite Sites in Plastidic Ferredoxin-NADP+ Reductase for the Interaction with NADP+" Antioxidants 11, no. 3: 537. https://doi.org/10.3390/antiox11030537
APA StylePérez-Domínguez, S., Caballero-Mancebo, S., Marcuello, C., Martínez-Júlvez, M., Medina, M., & Lostao, A. (2022). Nanomechanical Study of Enzyme: Coenzyme Complexes: Bipartite Sites in Plastidic Ferredoxin-NADP+ Reductase for the Interaction with NADP+. Antioxidants, 11(3), 537. https://doi.org/10.3390/antiox11030537