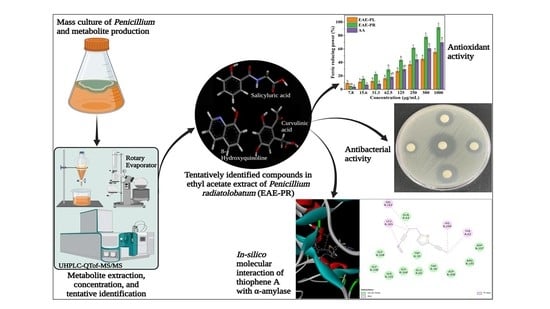
Comparative Analysis of the Antioxidant, Antidiabetic, Antibacterial, Cytoprotective Potential and Metabolite Profile of Two Endophytic Penicillium spp.
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Consumables
2.2. Fungal Culture Medium, Liquid Fermentation, and Metabolites Extraction
2.3. Assessment of Total Phenolics and Flavonoid Content
2.4. α-amylase and α-glucosidase Inhibition Assay
2.5. Antioxidant Assays
2.5.1. ABTS+ and DPPH Radical Scavenging Assay
2.5.2. Ferric-Reducing Power Assay
2.5.3. Determination of Peroxyl Radical Scavenging Capacity (PSC)
2.6. Antibacterial Susceptibility Test
Assessment of Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC)
2.7. Cell Viability Analysis (MTT Assay)
Cell Viability Analysis (Fluorescent Staining Assay)
2.8. Cytoprotective Activity against H2O2 Stress
2.9. Metabolite Profiling by UHPLC-Q-TOF-MS/MS
2.10. Molecular Docking Analysis
2.11. Statistical Analysis
3. Results and Discussion
3.1. Total Yield and Polyphenolics Quantification
3.2. α-amylase and α-glucosidase Inhibition Activities
3.3. Antioxidant Activities
3.4. Antibacterial Activity
3.5. Cell Viability Analysis
3.6. Cytoprotective Effect against H2O2 Stress
3.7. Metabolite Profiling by UHPLC-Q-TOF-MS/MS
3.8. Molecular Docking Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Noncommunicable Diseases Progress Monitor 2022; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- Yadav, M.; Yadav, A.; Yadav, J.P. In vitro antioxidant activity and total phenolic content of endophytic fungi isolated from Eugenia jambolana Lam. Asian Pac. J. Trop. Med. 2014, 7, S256–S261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anthony, K.P.; Saleh, M.A. Free radical scavenging and antioxidant activities of silymarin components. Antioxidants 2013, 2, 398–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheeseman, K.H.; Slater, T.F. An introduction to free radical biochemistry. Br. Med. Bull. 1993, 49, 481–493. [Google Scholar] [CrossRef]
- Ayoka, T.O.; Ezema, B.O.; Eze, C.N.; Nnadi, C.O. Antioxidants for the Prevention and Treatment of Non-communicable Diseases. J. Explor. Res. Pharmacol. 2022, 7, 178–188. [Google Scholar] [CrossRef]
- Sun, H.; Saeedi, P.; Karuranga, S.; Pinkepank, M.; Ogurtsova, K.; Duncan, B.B.; Stein, C.; Basit, A.; Chan, J.C.N.; Mbanya, J.C.; et al. IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract. 2022, 183, 109119. [Google Scholar] [CrossRef] [PubMed]
- Papoutsis, K.; Zhang, J.; Bowyer, M.C.; Brunton, N.; Gibney, E.R.; Lyng, J. Fruit, vegetables, and mushrooms for the preparation of extracts with α-amylase and α-glucosidase inhibition properties: A review. Food Chem. 2021, 338, 128119. [Google Scholar] [CrossRef]
- Sathiyaseelan, A.; Park, S.; Saravanakumar, K.; Mariadoss, A.V.A.; Wang, M.-H. Evaluation of phytochemicals, antioxidants, and antidiabetic efficacy of various solvent fractions of Gynura procumbens (Lour.) Merr. Process Biochem. 2021, 111, 51–62. [Google Scholar] [CrossRef]
- Mugaranja, K.P.; Kulal, A. Inhibition of alpha-glucosidase enzyme and antioxidant activity of the metabolites purified from an endophytic fungus of Simarouba glauca DC. S. Afr. J. Bot. 2022, 149, 134–144. [Google Scholar] [CrossRef]
- Saravanakumar, K.; Park, S.; Sathiyaseelan, A.; Kim, K.-N.; Cho, S.-H.; Mariadoss, A.V.A.; Wang, M.-H. Metabolite profiling of methanolic extract of Gardenia jaminoides by LC-MS/MS and GC-MS and its anti-diabetic, and anti-oxidant activities. Pharmaceuticals 2021, 14, 102. [Google Scholar] [CrossRef]
- Mariadoss, A.V.A.; Park, S.; Saravanakumar, K.; Sathiyaseelan, A.; Wang, M.-H. Ethyl acetate fraction of Helianthus tuberosus L. induces anti-diabetic, and wound-healing activities in insulin-resistant human liver cancer and mouse fibroblast cells. Antioxidants 2021, 10, 99. [Google Scholar] [CrossRef]
- Rai, N.; Gupta, P.; Keshri, P.K.; Verma, A.; Mishra, P.; Kumar, D.; Kumar, A.; Singh, S.K.; Gautam, V. Fungal Endophytes: An Accessible Source of Bioactive Compounds with Potential Anticancer Activity. Appl. Biochem. Biotechnol. 2022, 194, 3296–3319. [Google Scholar] [CrossRef] [PubMed]
- Naveen, K.V.; Sathiyaseelan, A.; Mariadoss, A.V.A.; Xiaowen, H.; Saravanakumar, K.; Wang, M.-H. Fabrication of mycogenic silver nanoparticles using endophytic fungal extract and their characterization, antibacterial and cytotoxic activities. Inorg. Chem. Commun. 2021, 128, 108575. [Google Scholar] [CrossRef]
- Liu, S.; Su, M.; Song, S.-J.; Jung, J.H. Marine-Derived Penicillium Species as Producers of Cytotoxic Metabolites. Mar. Drugs 2017, 15, 329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toghueo, R.M.K.; Boyom, F.F. Endophytic Penicillium species and their agricultural, biotechnological, and pharmaceutical applications. 3 Biotech 2020, 10, 107. [Google Scholar] [CrossRef] [PubMed]
- Ikuta, K.S.; Swetschinski, L.R.; Robles Aguilar, G.; Sharara, F.; Mestrovic, T.; Gray, A.P.; Davis Weaver, N.; Wool, E.E.; Han, C.; Gershberg Hayoon, A.; et al. Global mortality associated with 33 bacterial pathogens in 2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2022, 400, 2221–2248. [Google Scholar] [CrossRef]
- Oliver, S.P. Foodborne Pathogens and Disease Special Issue on the National and International PulseNet Network. Foodborne Pathog. Dis. 2019, 16, 439–440. [Google Scholar] [CrossRef]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States—Major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- Cueva, C.; Moreno-Arribas, M.V.; Martín-Álvarez, P.J.; Bills, G.; Vicente, M.F.; Basilio, A.; Rivas, C.L.; Requena, T.; Rodríguez, J.M.; Bartolomé, B. Antimicrobial activity of phenolic acids against commensal, probiotic and pathogenic bacteria. Res. Microbiol. 2010, 161, 372–382. [Google Scholar] [CrossRef]
- Tang, Z.; Qin, Y.; Chen, W.; Zhao, Z.; Lin, W.; Xiao, Y.; Chen, H.; Liu, Y.; Chen, H.; Bu, T.; et al. Diversity, chemical constituents, and biological activities of endophytic fungi isolated from Ligusticum chuanxiong Hort. Front. Microbiol. 2021, 12, 771000. [Google Scholar] [CrossRef]
- George, T.K.; Joy, A.; Divya, K.; Jisha, M.S. In vitro and in silico docking studies of antibacterial compounds derived from endophytic Penicillium setosum. Microb. Pathog. 2019, 131, 87–97. [Google Scholar] [CrossRef]
- Nischitha, R.; Shivanna, M.B. Metabolite fingerprinting, in vitro antimicrobial and antioxidant activities and in-silico docking in Alloteropsis cimicina and its endophytic fungus Penicillium pinophilum. Mol. Biol. Rep. 2021, 48, 4021–4037. [Google Scholar] [CrossRef] [PubMed]
- Gunarto, L.; Adachi, K.; Senboku, T. Isolation and selection of indigenous Azospirillum spp. from a subtropical island, and effect of inoculation on growth of lowland rice under several levels of N application. Biol. Fertil. Soils 1999, 28, 129–135. [Google Scholar] [CrossRef]
- Sathiyaseelan, A.; Saravanakumar, K.; Mariadoss, A.V.A.; Kim, K.M.; Wang, M.-H. Antibacterial activity of ethyl acetate extract of endophytic fungus (Paraconiothyrium brasiliense) through targeting dihydropteroate synthase (DHPS). Process Biochem. 2021, 111, 27–35. [Google Scholar] [CrossRef]
- Gao, X.; Xue, Z.; Ma, Q.; Guo, Q.; Xing, L.; Santhanam, R.K.; Zhang, M.; Chen, H. Antioxidant and antihypertensive effects of garlic protein and its hydrolysates and the related mechanism. J. Food Biochem. 2020, 44, e13126. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, A.; Chelliah, R.; Banan-Mwine Daliri, E.; Sultan, G.; Madar, I.H.; Kim, N.; Shabbir, U.; Oh, D.-H. Antioxidant activities of novel peptides from Limosilactobacillus reuteri fermented brown rice: A combined in vitro and in silico study. Food Chem. 2023, 404, 134747. [Google Scholar] [CrossRef] [PubMed]
- Rai, Y.; Pathak, R.; Kumari, N.; Sah, D.K.; Pandey, S.; Kalra, N.; Soni, R.; Dwarakanath, B.S.; Bhatt, A.N. Mitochondrial biogenesis and metabolic hyperactivation limits the application of MTT assay in the estimation of radiation induced growth inhibition. Sci. Rep. 2018, 8, 1531. [Google Scholar] [CrossRef] [Green Version]
- Naveen, K.V.; Saravanakumar, K.; Sathiyaseelan, A.; Wang, M.-H. Preparation, characterization, and synergistic antibacterial activity of mycosynthesized, PEGylated CuO nanoparticles combined tetracycline hydrochloride. J. Drug Deliv. Sci. Technol. 2022, 76, 103826. [Google Scholar] [CrossRef]
- Tyagi, A.; Yeon, S.-J.; Daliri, E.B.-M.; Chen, X.; Chelliah, R.; Oh, D.-H. Untargeted metabolomics of Korean fermented Brown rice using UHPLC Q-TOF MS/MS reveal an abundance of potential dietary antioxidative and stress-reducing compounds. Antioxidants 2021, 10, 626. [Google Scholar] [CrossRef]
- Da Silva, M.H.R.; Cueva-Yesquén, L.G.; Júnior, S.B.; Garcia, V.L.; Sartoratto, A.; de Angelis, D.d.F.; de Angelis, D.A. Endophytic fungi from Passiflora incarnata: An antioxidant compound source. Arch. Microbiol. 2020, 202, 2779–2789. [Google Scholar] [CrossRef]
- Rumidatul, A.; Rahmawati, N.; Sunarya, S. Production of Secondary Metabolites and its Antibacterial and Antioxidant Activity During the Growth Period of Endophytic Fungi Isolated from Gall Rust Sengon Plants. Pharmacogn. J. 2021, 13, 325–331. [Google Scholar] [CrossRef]
- Lu, L.; Feng, Q.; Su, T.; Cheng, Y.; Huang, Z.; Huang, Q.; Liu, Z. Chapter 35—Pharmacoepigenetics of Chinese Herbal Components in Cancer. In Pharmacoepigenetics; Cacabelos, R., Ed.; Academic Press: Cambridge, MA, USA, 2019; Volume 10, pp. 859–869. [Google Scholar]
- Ali, S.; Khan, A.L.; Ali, L.; Rizvi, T.S.; Khan, S.A.; Hussain, J.; Hamayun, M.; Al-Harrasi, A. Enzyme inhibitory metabolites from endophytic Penicillium citrinum isolated from Boswellia sacra. Arch. Microbiol. 2017, 199, 691–700. [Google Scholar] [CrossRef] [PubMed]
- Lü, J.M.; Lin, P.H.; Yao, Q.; Chen, C. Chemical and molecular mechanisms of antioxidants: Experimental approaches and model systems. J. Cell. Mol. Med. 2010, 14, 840–860. [Google Scholar] [CrossRef] [PubMed]
- Platzer, M.; Kiese, S.; Herfellner, T.; Schweiggert-Weisz, U.; Miesbauer, O.; Eisner, P. Common Trends and Differences in Antioxidant Activity Analysis of Phenolic Substances Using Single Electron Transfer Based Assays. Molecules 2021, 26, 1244. [Google Scholar] [CrossRef] [PubMed]
- Hulikere, M.M.; Joshi, C.G.; Ananda, D.; Poyya, J.; Nivya, T. Antiangiogenic, wound healing and antioxidant activity of Cladosporium cladosporioides (Endophytic Fungus) isolated from seaweed (Sargassum wightii). Mycology 2016, 7, 203–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spiegel, M.; Kapusta, K.; Kołodziejczyk, W.; Saloni, J.; Żbikowska, B.; Hill, G.A.; Sroka, Z. Antioxidant activity of selected phenolic acids–ferric reducing antioxidant power assay and QSAR analysis of the structural features. Molecules 2020, 25, 3088. [Google Scholar] [CrossRef] [PubMed]
- Moloney, M.G. Natural Products as a Source for Novel Antibiotics. Trends Pharmacol. Sci. 2016, 37, 689–701. [Google Scholar] [CrossRef]
- Rempe, C.S.; Burris, K.P.; Lenaghan, S.C.; Stewart, C.N., Jr. The potential of systems biology to discover antibacterial mechanisms of plant phenolics. Front. Microbiol. 2017, 8, 422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapoor, G.; Saigal, S.; Elongavan, A. Action and resistance mechanisms of antibiotics: A guide for clinicians. J. Anaesthesiol. Clin. Pharmacol. 2017, 33, 300–305. [Google Scholar] [CrossRef]
- Pasqualetti, V.; Locato, V.; Fanali, C.; Mulinacci, N.; Cimini, S.; Morgia, A.M.; Pasqua, G.; de Gara, L. Comparison between In Vitro Chemical and Ex Vivo Biological Assays to Evaluate Antioxidant Capacity of Botanical Extracts. Antioxidants 2021, 10, 1136. [Google Scholar] [CrossRef]
- Butkeviciute, A.; Ramanauskiene, K.; Kurapkiene, V.; Janulis, V. Dermal Penetration Studies of Potential Phenolic Compounds Ex Vivo and Their Antioxidant Activity In Vitro. Plants 2022, 11, 1901. [Google Scholar] [CrossRef]
- Hardaway, C.M.; Badisa, R.B.; Soliman, K.F.A. Effect of ascorbic acid and hydrogen peroxide on mouse neuroblastoma cells. Mol. Med. Rep. 2012, 5, 1449–1452. [Google Scholar] [PubMed]
- Podmore, I.D.; Griffiths, H.R.; Herbert, K.E.; Mistry, N.; Mistry, P.; Lunec, J. Vitamin C exhibits pro-oxidant properties. Nature 1998, 392, 559. [Google Scholar] [CrossRef] [PubMed]
- Divate, R.D.; Wang, P.-M.; Wang, C.-C.; Chou, S.-T.; Chang, C.-T.; Chung, Y.-C. Protective effect of medicinal fungus Xylaria nigripes mycelia extracts against hydrogen peroxide-induced apoptosis in PC12 cells. Int. J. Immunopathol. Pharmacol. 2017, 30, 105–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nischitha, R.; Shivanna, M.B. Diversity and in silico docking of antibacterial potent compounds in endophytic fungus Chaetomium subaffine Sergeeva and host Heteropogon contortus (L.) P. Beauv. Process Biochem. 2022, 112, 124–138. [Google Scholar] [CrossRef]
- Cook, S.D. An Historical Review of Phenylacetic Acid. Plant Cell Physiol. 2019, 60, 243–254. [Google Scholar] [CrossRef]
- Nicoletti, R.; Fiorentino, A. Plant Bioactive Metabolites and Drugs Produced by Endophytic Fungi of Spermatophyta. Agriculture 2015, 5, 918–970. [Google Scholar] [CrossRef] [Green Version]
- Ikram, M.; Ali, N.; Jan, G.; Hamayun, M.; Jan, F.G.; Iqbal, A. Novel antimicrobial and antioxidative activity by endophytic Penicillium roqueforti and Trichoderma reesei isolated from Solanum surattense. Acta Physiol. Plant. 2019, 41, 164. [Google Scholar] [CrossRef]
- Ananda, D.; Chandrashekhar, G.J.; Sunil Kumar, R.; Heggodu, G.R.K.; Balenahalli Narasingappa, R. Antioxidant and Cytotoxic Potential of Endophytic Fungi Isolated from Medicinal Plant Tragia involucrata L. Pharmacogn. Res. 2018, 10, 188–194. [Google Scholar]
- Srinivasa, C.; Mellappa, G.; Patil, S.M.; Ramu, R.; Shreevatsa, B.; Dharmashekar, C.; Kollur, S.P.; Syed, A.; Shivamallu, C. Plants and endophytes—A partnership for the coumarin production through the microbial systems. Mycology 2022, 13, 243–256. [Google Scholar] [CrossRef]
- Brader, G.; Compant, S.; Mitter, B.; Trognitz, F.; Sessitsch, A. Metabolic potential of endophytic bacteria. Curr. Opin. Biotechnol. 2014, 27, 30–37. [Google Scholar] [CrossRef] [Green Version]
- El-Hawary, S.S.; Sayed, A.M.; Mohammed, R.; Hassan, H.M.; Zaki, M.A.; Rateb, M.E.; Mohammed, T.A.; Amin, E.; Abdelmohsen, U.R. Epigenetic Modifiers Induce Bioactive Phenolic Metabolites in the Marine-Derived Fungus Penicillium brevicompactum. Mar. Drugs 2018, 16, 253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hawas, U.W.; El-Halawany, A.M.; Ahmed, E.F. Hepatitis C virus NS3-NS4A protease inhibitors from the endophytic Penicillium chrysogenum isolated from the red alga Liagora viscida. Z. Nat. C 2013, 68, 355–366. [Google Scholar]
- Oh, M.; Park, S.; Kim, H.; Choi, G.J.; Kim, S.H. Application of UPLC-QTOF-MS Based Untargeted Metabolomics in Identification of Metabolites Induced in Pathogen-Infected Rice. Plants 2021, 10, 213. [Google Scholar] [CrossRef] [PubMed]
- Dadwal, V.; Joshi, R.; Gupta, M. A comparative metabolomic investigation in fruit sections of Citrus medica L. and Citrus maxima L. detecting potential bioactive metabolites using UHPLC-QTOF-IMS. Food Res. Int. 2022, 157, 111486. [Google Scholar] [CrossRef] [PubMed]
- Prabhavathi, H.; Dasegowda, K.R.; Renukananda, K.H.; Karunakar, P.; Lingaraju, K.; Raja Naika, H. Molecular docking and dynamic simulation to identify potential phytocompound inhibitors for EGFR and HER2 as anti-breast cancer agents. J. Biomol. Struct. Dyn. 2022, 40, 4713–4724. [Google Scholar] [CrossRef]







| Sample | Extract Yield (%) | Total Phenolic Content (mg of GAE/g DW) | Total Flavonoid Content (mg of QE/g DW) |
|---|---|---|---|
| EAE-PL | 0.41 ± 0.08 a | 17.89 ± 1.63 a | 3.31 ± 0.96 a |
| EAE-PR | 0.63 ± 0.05 b | 119.87 ± 3.74 b | 16.26 ± 1.95 b |
| Sample | Half-Maximal Inhibitory Concentration (IC50; µg/mL) | |||||
|---|---|---|---|---|---|---|
| α-amylase Inhibition | α-glucosidase Inhibition | ABTS+ Radical Scavenging | DPPH Radical Scavenging | Ferric-Reducing Power | Peroxyl Radical Scavenging | |
| EAE-PL | >1000 c | >1000 c | 59.5 ± 1.54 c | >1000 c | 733.3 ± 2.61 c | >1000 c |
| EAE-PR | 362.5 ± 2.50 b | 525 ± 1.38 b | 37.5 ± 3.46 a | 187.5 ± 3.92 a | 175.0 ± 1.46 a | 532.36 ± 2.52 b |
| AA | ND | ND | 49.25 ± 2.10 b | 241.6 ± 1.95 b | 326.5 ± 3.78 b | 521.86 ± 1.75 a |
| AC | 37.5 ± 2.3 a | 87.5 ± 2.58 a | ND | ND | ND | ND |
| Bacterial Strains | Zone of Inhibition (mm) | ||||
|---|---|---|---|---|---|
| EAE-PL (µg/mL) | EAE-PR (µg/mL) | Ery (100 µg/mL) | |||
| 500 | 1000 | 500 | 1000 | ||
| B. cereus | 9 ± 0.5 b | 11.5 ± 1.0 d | 14 ± 0.3 c | 19 ± 1.5 b | 22 ± 1.5 b |
| S. aureus | 0 | 8 ± 0.3 a | 11 ± 0.1 a | 18 ± 0.5 a | 22 ± 0.3 b |
| L. monocytogenes | 0 | 9 ± 0.2 b | 15 ± 0.5 d | 19 ± 1.5 b | 21 ± 1.0 a |
| E. coli | 6.5 ± 0.2 a | 11.5 ± 0.5 e | 17 ± 0.1 e | 19 ± 1.0 b | 22 ± 0.5 b |
| S. enterica | 0 | 10 ± 0.3 c | 13 ± 0.2 b | 18 ± 0.5 a | 21 ± 0.8 a |
| Bacterial Strains | MIC (µg/mL) | MBC (µg/mL) | ||
|---|---|---|---|---|
| EAE-PL | EAE-PR | EAE-PL | EAE-PR | |
| B. cereus | 62.5 a | 31.25 a | 500 a | 125 a |
| S. aureus | 500 d | 125 c | 1000 b | 500 c |
| L. monocytogenes | 500 d | 31.25 a | 1000 b | 125 a |
| E. coli | 125 b | 62.5 b | 500 a | 250 b |
| S. enterica | 250 c | 62.5 b | 1000 b | 250 b |
| S.No. | RT (min) | Tentatively Identified Compounds | Formula | Molecular Weight | Found at Mass | Area | Adduct/Charge |
|---|---|---|---|---|---|---|---|
| 1. | 1.23 | Nicotinic Acid | C6H5NO2 | 123.03215 | 124.0388 | 2.2 × 106 | [M+H]+ |
| 2. | 1.64 | Succinic anhydride | C4H4O3 | 100.01648 | 101.0233 | 2.4 × 105 | [M+H]+ |
| 3. | 2.03 | Myrcene | C10H16 | 136.12567 | 137.1324 | 3.0 × 104 | [M+H]+ |
| 4. | 3.33 | 2-Methoxyresorcinol | C7H8O3 | 140.04776 | 141.0544 | 3.4 × 106 | [M+H]+ |
| 5. | 5.17 | Sorbic Acid | C6H8O2 | 112.05289 | 113.0597 | 1.1 × 106 | [M+H]+ |
| 6. | 6.42 | Dihydro-beta-tubaic acid | C13H16O4 | 236.10549 | 237.1124 | 4.6 × 105 | [M+H]+ |
| 7. | 7.35 | Trigonelline | C7H7NO2 | 137.04816 | 138.055 | 5.7 × 105 | [M+H]+ |
| 8. | 9.12 | Sinapaldehyde | C11H12O4 | 208.07402 | 209.0809 | 4.5 × 105 | [M+H]+ |
| 9. | 9.15 | Radicinin | C12H12O5 | 236.06843 | 237.0751 | 1.1 × 106 | [M+H]+ |
| 10. | 9.36 | Acetophenone | C8H8O | 120.05784 | 121.0645 | 4.3 × 106 | [M+H]+ |
| 11. | 10.91 | Vanillyl alcohol | C8H10O3 | 154.06361 | 155.0703 | 3.9 × 106 | [M+H]+ |
| 12. | 11.41 | p-Hydroxyphenylacetic acid | C8H8O3 | 152.04762 | 153.0543 | 2.8 × 107 | [M+H]+ |
| 13. | 11.51 | Penicillic acid | C8H10O4 | 170.05863 | 169.051 | 1.7 × 105 | [M-H]- |
| 14. | 13.08 | 3-Phenylpropionic acid | C9H10O2 | 150.06885 | 151.0755 | 7.1 × 105 | [M+H]+ |
| 15. | 13.36 | Phenylacetic acid | C8H8O2 | 136.05302 | 137.0598 | 5.3 × 105 | [M+H]+ |
| 16. | 14.47 | Valacyclovir | C13H20N6O4 | 324.15505 | 325.1615 | 7.2 × 106 | [M+H]+ |
| 17. | 15.85 | Ferulic acid | C10H10O4 | 194.05872 | 195.0655 | 1.2 × 106 | [M+H]+ |
| 18. | 16.54 | Eugenol | C10H12O2 | 164.08434 | 165.091 | 1.0 × 106 | [M+H]+ |
| 19. | 16.61 | Epicatechin | C15H14O6 | 290.07957 | 291.0866 | 1.2 × 106 | [M+H]+ |
| 20. | 16.77 | Butylphthalide | C12H14O2 | 190.10013 | 191.1068 | 1.7 × 106 | [M+H]+ |
| S.No. | RT (min) | Tentatively Identified Compounds | Formula | Molecular Weight | Found at Mass | Area | Adduct/Charge |
|---|---|---|---|---|---|---|---|
| 1. | 0.88 | Salicyluric acid | C9H9NO4 | 195.05402 | 196.0607 | 1.9 × 105 | [M+H]+ |
| 2. | 2.4 | Limonene | C10H16 | 136.12562 | 137.1325 | 1.1 × 105 | [M+H]+ |
| 3. | 2.59 | Thiophene A | C13H8S | 196.03571 | 197.0424 | 4.8 × 105 | [M+H]+ |
| 4. | 3.44 | Zeatin | C10H13N5O | 219.11283 | 220.1194 | 9.0 × 105 | [M+H]+ |
| 5. | 5.17 | Sorbic Acid | C6H8O2 | 112.05289 | 113.0597 | 2.5 × 105 | [M+H]+ |
| 6. | 6.82 | 3-Indoleacetic acid | C10H9NO2 | 175.06414 | 176.0708 | 6.2 × 105 | [M-H]- |
| 7. | 7.93 | 8-Hydroxyquinoline | C9H7NO | 145.05328 | 146.06 | 4.7 × 105 | [M+H]+ |
| 8. | 8.13 | Maculosin | C14H16N2O3 | 260.11727 | 261.1238 | 1.1 × 106 | [M+H]+ |
| 9. | 8.58 | Dehydroacetic acid | C8H8O4 | 168.04297 | 169.0497 | 9.0 × 105 | [M+H]+ |
| 10. | 8.68 | Esculetin | C9H6O4 | 178.02731 | 179.0342 | 3.4 × 105 | [M+H]+ |
| 11. | 9.12 | Sinapaldehyde | C11H12O4 | 208.07402 | 209.081 | 4.9 × 105 | [M+H]+ |
| 12. | 9.81 | Foetisulfide A | C8H16OS3 | 224.03749 | 225.0438 | 5.8 × 103 | [M+H]+ |
| 13. | 10.93 | Haematommic acid | C9H8O5 | 196.03785 | 197.0447 | 3.8 × 105 | [M+H]+ |
| 14. | 11.08 | p-Coumaric acid | C9H8O3 | 164.04811 | 165.0547 | 1.4 × 106 | [M+H]+ |
| 15. | 12.28 | Vanillin | C8H8O3 | 152.04761 | 153.0542 | 2.6 × 107 | [M+H]+ |
| 16. | 12.73 | Scopoletin | C10H8O4 | 192.04292 | 193.0495 | 1.3 × 107 | [M+H]+ |
| 17. | 12.76 | Curvulinic acid | C10H10O5 | 210.05359 | 209.046 | 1.3 × 106 | [M-H]- |
| 18. | 12.89 | Sinapic acid | C11H12O5 | 224.06794 | 225.0753 | 1.2 × 107 | [M+H]+ |
| 19. | 14.6 | Dehydrochorismic Acid | C10H8O6 | 224.03286 | 225.0395 | 4.5 × 105 | [M+H]+ |
| 20. | 14.98 | Harmine | C13H12N2O | 212.09586 | 213.1027 | 3.4 × 105 | [M+H]+ |
| 21. | 15.01 | 4-Methoxycinnamic acid | C10H10O3 | 178.06369 | 179.0704 | 1.3 × 106 | [M+H]+ |
| 22. | 15.84 | Ethylparaben | C9H10O3 | 166.06370 | 167.0705 | 1.9 × 106 | [M+H]+ |
| 23. | 15.85 | Ferulic acid | C10H10O4 | 194.05872 | 195.0646 | 2.6 × 107 | [M+H]+ |
| 24. | 16.05 | Fraxetin | C10H8O5 | 208.03726 | 209.0439 | 8.7 × 106 | [M+H]+ |
| 25. | 16.41 | Isofraxidin | C11H10O5 | 222.05344 | 221.0459 | 4.0 × 105 | [M-H]- |
| 26. | 16.54 | Phenylacetic acid | C8H8O2 | 136.05302 | 137.0598 | 1.1 × 106 | [M+H]+ |
| Compounds | α-amylase | α-glucosidase | NADPH Oxidase | D-alanine D-alanine Ligase |
|---|---|---|---|---|
| 3-Indoleacetic acid | −8.77 | −9.60 | −8.88 | −9.11 |
| 4-Methoxycinnamic acid | −8.28 | −8.06 | −8.80 | −9.17 |
| 8-Hydroxyquinoline | −8.35 | −8.85 | −8.47 | −9.04 |
| Curvulinic acid | −8.07 | −8.63 | −8.59 | −9.04 |
| Dehydroacetic acid | −6.54 | −6.99 | −7.33 | −6.87 |
| Dehydrochorismic Acid | −7.59 | −8.39 | −8.05 | −8.51 |
| Esculetin | −8.22 | −8.14 | −8.15 | −8.39 |
| Ethylparaben | −8.50 | −8.43 | −8.33 | −8.79 |
| Ferulic acid | −7.85 | −8.65 | −8.53 | −8.93 |
| Foetisulfide A | −8.31 | −9.73 | −8.99 | −10.39 |
| Fraxetin | −7.95 | −7.70 | −7.80 | −8.13 |
| Haematommic acid | −9.00 | −8.52 | −7.86 | −8.36 |
| Harmine | −8.14 | −8.06 | −8.27 | −8.08 |
| Isofraxidin | −7.99 | −8.03 | −8.10 | −7.72 |
| Limonene | −8.78 | −10.17 | −9.65 | −12.40 |
| Maculosin | −8.75 | −8.83 | −9.28 | −8.75 |
| p-Coumaric acid | −8.51 | −9.75 | −9.06 | −9.25 |
| Phenylacetic acid | −8.75 | −9.99 | −8.87 | −10.61 |
| Salicyluric acid | −8.34 | −8.63 | −8.70 | −9.26 |
| Scopoletin | −7.60 | −7.28 | −8.27 | −8.05 |
| Sinapaldehyde | −7.87 | −8.39 | −8.80 | −8.60 |
| Sinapic acid | −8.54 | −7.84 | −8.59 | −8.45 |
| Sorbic Acid | −7.99 | −9.57 | −7.73 | −9.09 |
| Thiophene A | −9.75 | −11.74 | −10.96 | −12.06 |
| Vanillin | −7.29 | −8.09 | −7.81 | −8.12 |
| Zeatin | −7.85 | −7.91 | −7.64 | −7.87 |
| Acarbose | −6.81 | −7.52 | ND | ND |
| Ascorbic acid | ND | ND | −7.47 | ND |
| Erythronolide A (Ery) | ND | ND | ND | −9.07 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naveen, K.V.; Saravanakumar, K.; Sathiyaseelan, A.; Wang, M.-H. Comparative Analysis of the Antioxidant, Antidiabetic, Antibacterial, Cytoprotective Potential and Metabolite Profile of Two Endophytic Penicillium spp. Antioxidants 2023, 12, 248. https://doi.org/10.3390/antiox12020248
Naveen KV, Saravanakumar K, Sathiyaseelan A, Wang M-H. Comparative Analysis of the Antioxidant, Antidiabetic, Antibacterial, Cytoprotective Potential and Metabolite Profile of Two Endophytic Penicillium spp. Antioxidants. 2023; 12(2):248. https://doi.org/10.3390/antiox12020248
Chicago/Turabian StyleNaveen, Kumar Vishven, Kandasamy Saravanakumar, Anbazhagan Sathiyaseelan, and Myeong-Hyeon Wang. 2023. "Comparative Analysis of the Antioxidant, Antidiabetic, Antibacterial, Cytoprotective Potential and Metabolite Profile of Two Endophytic Penicillium spp." Antioxidants 12, no. 2: 248. https://doi.org/10.3390/antiox12020248
APA StyleNaveen, K. V., Saravanakumar, K., Sathiyaseelan, A., & Wang, M. -H. (2023). Comparative Analysis of the Antioxidant, Antidiabetic, Antibacterial, Cytoprotective Potential and Metabolite Profile of Two Endophytic Penicillium spp. Antioxidants, 12(2), 248. https://doi.org/10.3390/antiox12020248