Silver Nanoparticles Induce Mitochondrial Protein Oxidation in Lung Cells Impacting Cell Cycle and Proliferation
Abstract
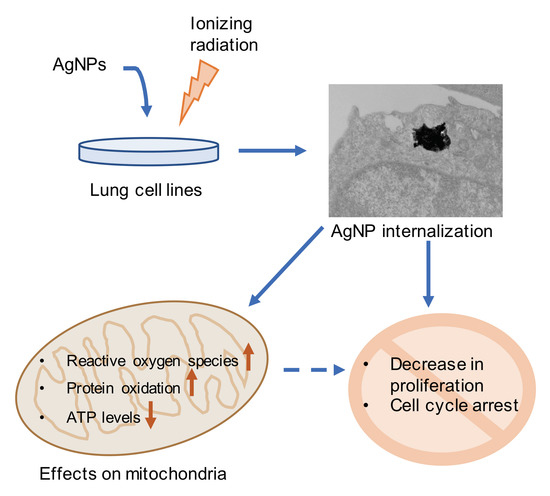
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Treatment with AgNPs and Ionizing Radiation
2.3. Analysis of Mitochondrial ROS and Protein Oxidation
2.4. Transmission Electron Microscopy (TEM)
2.5. Flow Cytometry (Mitochondrial Content, Mitochondrial Protein Oxidation, Mitochondrial ROS and Cell Cycle Analysis)
2.6. Cell Proliferation and ATP Assay
2.7. Mitochondrial Respirometry Analysis
3. Results
3.1. Cell Proliferation
3.2. Cell Cycle
3.3. Mitochondrial Reactive Oxygen Species (ROS)
3.4. Mitochondrial Protein Oxidation
3.5. Mitochondrial Function
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Akter, M.; Sikder, M.T.; Rahman, M.M.; Ullah, A.; Hossain, K.F.B.; Banik, S.; Hosokawa, T.; Saito, T.; Kurasaki, M. A systematic review on silver nanoparticles-induced cytotoxicity: Physicochemical properties and perspectives. J. Adv. Res. 2018, 9, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Nymark, P.; Kohonen, P.; Hongisto, V.; Grafstrom, R.C. Toxic and Genomic Influences of Inhaled Nanomaterials as a Basis for Predicting Adverse Outcome. Ann. Am. Thorac. Soc. 2018, 15, S91–S97. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.J.; Kantoff, P.W.; Wooster, R.; Farokhzad, O.C. Cancer nanomedicine: Progress, challenges and opportunities. Nat. Rev. Cancer 2017, 17, 20–37. [Google Scholar] [CrossRef] [PubMed]
- Burdusel, A.C.; Gherasim, O.; Grumezescu, A.M.; Mogoanta, L.; Ficai, A.; Andronescu, E. Biomedical Applications of Silver Nanoparticles: An Up-to-Date Overview. Nanomaterials 2018, 8, 681. [Google Scholar] [CrossRef]
- Jeong, J.K.; Gurunathan, S.; Kang, M.H.; Han, J.W.; Das, J.; Choi, Y.J.; Kwon, D.N.; Cho, S.G.; Park, C.; Seo, H.G.; et al. Hypoxia-mediated autophagic flux inhibits silver nanoparticle-triggered apoptosis in human lung cancer cells. Sci. Rep. 2016, 6, 21688. [Google Scholar] [CrossRef]
- McGillicuddy, E.; Murray, I.; Kavanagh, S.; Morrison, L.; Fogarty, A.; Cormican, M.; Dockery, P.; Prendergast, M.; Rowan, N.; Morris, D. Silver nanoparticles in the environment: Sources, detection and ecotoxicology. Sci. Total Environ. 2017, 575, 231–246. [Google Scholar] [CrossRef]
- Chen, X.; Schluesener, H.J. Nanosilver: A nanoproduct in medical application. Toxicol. Lett. 2008, 176, 1–12. [Google Scholar] [CrossRef]
- Dos Santos, C.A.; Seckler, M.M.; Ingle, A.P.; Gupta, I.; Galdiero, S.; Galdiero, M.; Gade, A.; Rai, M. Silver nanoparticles: Therapeutical uses, toxicity, and safety issues. J. Pharm. Sci. 2014, 103, 1931–1944. [Google Scholar] [CrossRef]
- Liu, P.D.; Huang, Z.H.; Chen, Z.W.; Xu, R.Z.; Wu, H.; Zang, F.C.; Wang, C.L.; Gu, N. Silver nanoparticles: A novel radiation sensitizer for glioma? Nanoscale 2013, 5, 11829–11836. [Google Scholar] [CrossRef]
- Locatelli, E.; Naddaka, M.; Uboldi, C.; Loudos, G.; Fragogeorgi, E.; Molinari, V.; Pucci, A.; Tsotakos, T.; Psimadas, D.; Ponti, J.; et al. Targeted delivery of silver nanoparticles and alisertib: In vitro and in vivo synergistic effect against glioblastoma. Nanomedicine 2014, 9, 839–849. [Google Scholar] [CrossRef]
- Miura, N.; Shinohara, Y. Cytotoxic effect and apoptosis induction by silver nanoparticles in HeLa cells. Biochem. Biophys. Res. Commun. 2009, 390, 733–737. [Google Scholar] [CrossRef] [PubMed]
- Kawata, K.; Osawa, M.; Okabe, S. In vitro toxicity of silver nanoparticles at noncytotoxic doses to HepG2 human hepatoma cells. Environ. Sci. Technol. 2009, 43, 6046–6051. [Google Scholar] [CrossRef] [PubMed]
- Sanpui, P.; Chattopadhyay, A.; Ghosh, S.S. Induction of Apoptosis in Cancer Cells at Low Silver Nanoparticle Concentrations using Chitosan Nanocarrier. Acs Appl. Mater. Interfaces 2011, 3, 218–228. [Google Scholar] [CrossRef] [PubMed]
- Beer, C.; Foldbjerg, R.; Hayashi, Y.; Sutherland, D.S.; Autrup, H. Toxicity of silver nanoparticles-Nanoparticle or silver ion? Toxicol. Lett. 2012, 208, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Zielinska, E.; Zauszkiewicz-Pawlak, A.; Wojcik, M.; Inkielewicz-Stepniak, I. Silver nanoparticles of different sizes induce a mixed type of programmed cell death in human pancreatic ductal adenocarcinoma. Oncotarget 2018, 9, 4675–4697. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.H.; Zhao, Y.X.; Guo, Q.Q.; Wang, Z.; Wang, H.Y.; Yang, Y.X.; Huang, Y.Z. TAT-modified nanosilver for combating multidrug-resistant cancer. Biomaterials 2012, 33, 6155–6161. [Google Scholar] [CrossRef] [PubMed]
- Fahrenholtz, C.D.; Swanner, J.; Ramirez-Perez, M.; Singh, R.N. Heterogeneous Responses of Ovarian Cancer Cells to Silver Nanoparticles as a Single Agent and in Combination with Cisplatin. J. Nanomater. 2017, 2017, 5107485. [Google Scholar] [CrossRef]
- Guo, D.; Zhu, L.; Huang, Z.; Zhou, H.; Ge, Y.; Ma, W.; Wu, J.; Zhang, X.; Zhou, X.; Zhang, Y.; et al. Anti-leukemia activity of PVP-coated silver nanoparticles via generation of reactive oxygen species and release of silver ions. Biomaterials 2013, 34, 7884–7894. [Google Scholar] [CrossRef]
- Guo, D.W.; Zhao, Y.; Zhang, Y.; Wang, Q.; Huang, Z.H.; Ding, Q.; Guo, Z.R.; Zhou, X.F.; Zhu, L.Y.; Gu, N. The Cellular Uptake and Cytotoxic Effect of Silver Nanoparticles on Chronic Myeloid Leukemia Cells. J. Biomed. Nanotechnol. 2014, 10, 669–678. [Google Scholar] [CrossRef]
- Carlson, C.; Hussain, S.M.; Schrand, A.M.; Braydich-Stolle, L.K.; Hess, K.L.; Jones, R.L.; Schlager, J.J. Unique cellular interaction of silver nanoparticles: Size-dependent generation of reactive oxygen species. J. Phys. Chem. B 2008, 112, 13608–13619. [Google Scholar] [CrossRef]
- Foldbjerg, R.; Dang, D.A.; Autrup, H. Cytotoxicity and genotoxicity of silver nanoparticles in the human lung cancer cell line, A549. Arch. Toxicol. 2011, 85, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Cheng, F.Y.; Chiu, H.W.; Tsai, J.C.; Fang, C.Y.; Chen, C.W.; Wang, Y.J. Cytotoxicity, oxidative stress, apoptosis and the autophagic effects of silver nanoparticles in mouse embryonic fibroblasts. Biomaterials 2014, 35, 4706–4715. [Google Scholar] [CrossRef] [PubMed]
- AshaRani, P.V.; Low Kah Mun, G.; Hande, M.P.; Valiyaveettil, S. Cytotoxicity and genotoxicity of silver nanoparticles in human cells. ACS Nano 2009, 3, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Guo, D.; Sun, L.; Huang, Z.; Zhang, X.; Ma, W.; Wu, J.; Xiao, L.; Zhao, Y.; Gu, N. Activation of autophagy by elevated reactive oxygen species rather than released silver ions promotes cytotoxicity of polyvinylpyrrolidone-coated silver nanoparticles in hematopoietic cells. Nanoscale 2017, 9, 5489–5498. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Buckley, A.; Marczylo, T.; Seiffert, J.; Romer, I.; Warren, J.; Hodgson, A.; Chung, K.F.; Gant, T.W.; Smith, R.; et al. The small airway epithelium as a target for the adverse pulmonary effects of silver nanoparticle inhalation. Nanotoxicology 2018, 6, 539–553. [Google Scholar] [CrossRef]
- Braakhuis, H.M.; Gosens, I.; Krystek, P.; Boere, J.A.; Cassee, F.R.; Fokkens, P.H.; Post, J.A.; van Loveren, H.; Park, M.V. Particle size dependent deposition and pulmonary inflammation after short-term inhalation of silver nanoparticles. Part Fibre Toxicol. 2014, 11, 49. [Google Scholar] [CrossRef]
- Seiffert, J.; Buckley, A.; Leo, B.; Martin, N.G.; Zhu, J.; Dai, R.; Hussain, F.; Guo, C.; Warren, J.; Hodgson, A.; et al. Pulmonary effects of inhalation of spark-generated silver nanoparticles in Brown-Norway and Sprague-Dawley rats. Respir. Res. 2016, 17, 85. [Google Scholar] [CrossRef]
- Tyrrell, D.J.; Bharadwaj, M.S.; Van Horn, C.G.; Kritchevsky, S.B.; Nicklas, B.J.; Molina, A.J. Respirometric Profiling of Muscle Mitochondria and Blood Cells Are Associated With Differences in Gait Speed Among Community-Dwelling Older Adults. J. Gerontol. A Biol. Sci. Med. Sci. 2015, 70, 1394–1399. [Google Scholar] [CrossRef]
- Bharadwaj, M.S.; Tyrrell, D.J.; Leng, I.; Demons, J.L.; Lyles, M.F.; Carr, J.J.; Nicklas, B.J.; Molina, A.J. Relationships between mitochondrial content and bioenergetics with obesity, body composition and fat distribution in healthy older adults. BMC Obes. 2015, 2, 40. [Google Scholar] [CrossRef]
- Wallace, D.C. Mitochondria and cancer. Nat. Rev.Cancer 2012, 12, 685–698. [Google Scholar] [CrossRef]
- Arun, S.; Liu, L.; Donmez, G. Mitochondrial Biology and Neurological Diseases. Curr. Neuropharmacol. 2016, 14, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Sadoshima, J. Molecular mechanisms of mitochondrial autophagy/mitophagy in the heart. Circ. Res. 2015, 116, 1477–1490. [Google Scholar] [CrossRef] [PubMed]
- Requejo, R.; Chouchani, E.T.; Hurd, T.R.; Menger, K.E.; Hampton, M.B.; Murphy, M.P. Measuring mitochondrial protein thiol redox state. Methods Enzymol. 2010, 474, 123–147. [Google Scholar] [CrossRef] [PubMed]
- Caito, S.W.; Aschner, M. Mitochondrial Redox Dysfunction and Environmental Exposures. Antioxid. Redox Signal. 2015, 23, 578–595. [Google Scholar] [CrossRef]
- Devarie-Baez, N.O.; Silva Lopez, E.I.; Furdui, C.M. Biological chemistry and functionality of protein sulfenic acids and related thiol modifications. Free Radic. Res. 2016, 50, 172–194. [Google Scholar] [CrossRef] [Green Version]
- Sena, L.A.; Chandel, N.S. Physiological Roles of Mitochondrial Reactive Oxygen Species. Mol. Cell 2012, 48, 158–167. [Google Scholar] [CrossRef] [Green Version]
- Holmila, R.J.; Vance, S.A.; Chen, X.; Wu, H.; Shukla, K.; Bharadwaj, M.S.; Mims, J.; Wary, Z.; Marrs, G.; Singh, R.; et al. Mitochondria-targeted Probes for Imaging Protein Sulfenylation. Sci. Rep. 2018, 8, 6635. [Google Scholar] [CrossRef] [Green Version]
- Kleinauskas, A.; Rocha, S.; Sahu, S.; Sun, Y.P.; Juzenas, P. Carbon-core silver-shell nanodots as sensitizers for phototherapy and radiotherapy. Nanotechnology 2013, 24, 325103. [Google Scholar] [CrossRef]
- Swanner, J.; Mims, J.; Carroll, D.L.; Akman, S.A.; Furdui, C.M.; Torti, S.V.; Singh, R.N. Differential cytotoxic and radiosensitizing effects of silver nanoparticles on triple-negative breast cancer and non-triple-negative breast cells. Int. J. Nanomed. 2015, 10, 3937–3953. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, K.C.; Seligy, V.L.; Massarsky, A.; Moon, T.W.; Rippstein, P.; Tan, J.; Tayabali, A.F. Comparison of toxicity of uncoated and coated silver nanoparticles. J. Phys. Conf. Ser. 2013, 429, 012025. [Google Scholar] [CrossRef] [Green Version]
- Swanner, J.; Fahrenholtz, C.D.; Tenvooren, I.; Bernish, B.W.; Sears, J.J.; Hooker, A.; Furdui, C.M.; Alli, E.; Li, W.; Donati, G.L.; et al. Silver nanoparticles selectively treat triple-negative breast cancer cells without affecting non-malignant breast epithelial cells in vitro and in vivo. FASEB BioAdv. 2019, 1, 639–660. [Google Scholar] [CrossRef]
- Yamamori, T.; Yasui, H.; Yamazumi, M.; Wada, Y.; Nakamura, Y.; Nakamura, H.; Inanami, O. Ionizing radiation induces mitochondrial reactive oxygen species production accompanied by upregulation of mitochondrial electron transport chain function and mitochondrial content under control of the cell cycle checkpoint. Free Radic. Biol. Med. 2012, 53, 260–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurunathan, S.; Qasim, M.; Park, C.; Yoo, H.; Kim, J.H.; Hong, K. Cytotoxic Potential and Molecular Pathway Analysis of Silver Nanoparticles in Human Colon Cancer Cells HCT116. Int. J. Mol. Sci. 2018, 19, 2269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cameron, S.J.; Hosseinian, F.; Willmore, W.G. A Current Overview of the Biological and Cellular Effects of Nanosilver. Int. J. Mol. Sci. 2018, 19, 30. [Google Scholar] [CrossRef] [Green Version]
- Jezek, J.; Cooper, K.F.; Strich, R. Reactive Oxygen Species and Mitochondrial Dynamics: The Yin and Yang of Mitochondrial Dysfunction and Cancer Progression. Antioxidants 2018, 7, 13. [Google Scholar] [CrossRef]
- Wu, H.; Lin, J.; Liu, P.; Huang, Z.; Zhao, P.; Jin, H.; Ma, J.; Wen, L.; Gu, N. Reactive oxygen species acts as executor in radiation enhancement and autophagy inducing by AgNPs. Biomaterials 2016, 101, 1–9. [Google Scholar] [CrossRef]
- Wu, H.; Lin, J.; Liu, P.; Huang, Z.; Zhao, P.; Jin, H.; Wang, C.; Wen, L.; Gu, N. Is the autophagy a friend or foe in the silver nanoparticles associated radiotherapy for glioma? Biomaterials 2015, 62, 47–57. [Google Scholar] [CrossRef]
- Zheng, Q.; Yang, H.; Wei, J.; Tong, J.L.; Shu, Y.Q. The role and mechanisms of nanoparticles to enhance radiosensitivity in hepatocellular cell. Biomed. Pharmacother. 2013, 67, 569–575. [Google Scholar] [CrossRef]
- Che, B.; Luo, Q.; Zhai, B.; Fan, G.; Liu, Z.; Cheng, K.; Xin, L. Cytotoxicity and genotoxicity of nanosilver in stable GADD45alpha promoter-driven luciferase reporter HepG2 and A549 cells. Environ. Toxicol. 2017, 32, 2203–2211. [Google Scholar] [CrossRef]
- Rosario, F.; Hoet, P.; Santos, C.; Oliveira, H. Death and cell cycle progression are differently conditioned by the AgNP size in osteoblast-like cells. Toxicology 2016, 368, 103–115. [Google Scholar] [CrossRef]
- Eom, H.J.; Choi, J. p38 MAPK activation, DNA damage, cell cycle arrest and apoptosis as mechanisms of toxicity of silver nanoparticles in Jurkat T cells. Environ. Sci. Technol. 2010, 44, 8337–8342. [Google Scholar] [CrossRef] [PubMed]
- Loutfy, S.A.; Al-Ansary, N.A.; Abdel-Ghani, N.T.; Hamed, A.R.; Mohamed, M.B.; Craik, J.D.; Eldin, T.A.; Abdellah, A.M.; Hussein, Y.; Hasanin, M.T.; et al. Anti-proliferative Activities of Metallic Nanoparticles in an in Vitro Breast Cancer Model. Asian Pac. J. Cancer Prev. 2015, 16, 6039–6046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawamura, K.; Qi, F.; Kobayashi, J. Potential relationship between the biological effects of low-dose irradiation and mitochondrial ROS production. J. Radiat. Res. 2018, 59, ii91–ii97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kam, W.W.; Banati, R.B. Effects of ionizing radiation on mitochondria. Free Radic. Biol. Med. 2013, 65, 607–619. [Google Scholar] [CrossRef]
- Gorrini, C.; Harris, I.S.; Mak, T.W. Modulation of oxidative stress as an anticancer strategy. Nat. Rev. Drug Discov. 2013, 12, 931–947. [Google Scholar] [CrossRef]
- Menon, S.G.; Goswami, P.C. A redox cycle within the cell cycle: Ring in the old with the new. Oncogene 2007, 26, 1101–1109. [Google Scholar] [CrossRef] [Green Version]
- Sarsour, E.H.; Kumar, M.G.; Chaudhuri, L.; Kalen, A.L.; Goswami, P.C. Redox control of the cell cycle in health and disease. Antioxid. Redox Signal. 2009, 11, 2985–3011. [Google Scholar] [CrossRef]
- Ma, W.; Jing, L.; Valladares, A.; Mehta, S.L.; Wang, Z.; Li, P.A.; Bang, J.J. Silver nanoparticle exposure induced mitochondrial stress, caspase-3 activation and cell death: Amelioration by sodium selenite. Int. J. Biol. Sci. 2015, 11, 860–867. [Google Scholar] [CrossRef] [Green Version]
- Maurer, L.L.; Meyer, J.N. A systematic review of evidence for silver nanoparticle-induced mitochondrial toxicity. Environ. Sci. Nano 2016, 3, 311–322. [Google Scholar] [CrossRef]
- Takenaka, S.; Karg, E.; Roth, C.; Schulz, H.; Ziesenis, A.; Heinzmann, U.; Schramel, P.; Heyder, J. Pulmonary and systemic distribution of inhaled ultrafine silver particles in rats. Environ. Health Perspect. 2001, 109 (Suppl. 4), 547–551. [Google Scholar] [CrossRef] [Green Version]
- Marchi, E.; Vargas, F.S.; Teixeira, L.R.; Acencio, M.M.; Antonangelo, L.; Light, R.W. Intrapleural low-dose silver nitrate elicits more pleural inflammation and less systemic inflammation than low-dose talc. Chest 2005, 128, 1798–1804. [Google Scholar] [CrossRef] [PubMed]
- Arsenopoulou, Z.V.; Taitzoglou, I.A.; Molyvdas, P.A.; Gourgoulianis, K.I.; Hatzoglou, C.; Zarogiannis, S.G. Silver nanoparticles alter the permeability of sheep pleura and of sheep and human pleural mesothelial cell monolayers. Environ. Toxicol. Pharmacol. 2017, 50, 212–215. [Google Scholar] [CrossRef] [PubMed]







© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Holmila, R.J.; Vance, S.A.; King, S.B.; Tsang, A.W.; Singh, R.; Furdui, C.M. Silver Nanoparticles Induce Mitochondrial Protein Oxidation in Lung Cells Impacting Cell Cycle and Proliferation. Antioxidants 2019, 8, 552. https://doi.org/10.3390/antiox8110552
Holmila RJ, Vance SA, King SB, Tsang AW, Singh R, Furdui CM. Silver Nanoparticles Induce Mitochondrial Protein Oxidation in Lung Cells Impacting Cell Cycle and Proliferation. Antioxidants. 2019; 8(11):552. https://doi.org/10.3390/antiox8110552
Chicago/Turabian StyleHolmila, Reetta J., Stephen A. Vance, Stephen Bruce King, Allen W. Tsang, Ravi Singh, and Cristina M. Furdui. 2019. "Silver Nanoparticles Induce Mitochondrial Protein Oxidation in Lung Cells Impacting Cell Cycle and Proliferation" Antioxidants 8, no. 11: 552. https://doi.org/10.3390/antiox8110552
APA StyleHolmila, R. J., Vance, S. A., King, S. B., Tsang, A. W., Singh, R., & Furdui, C. M. (2019). Silver Nanoparticles Induce Mitochondrial Protein Oxidation in Lung Cells Impacting Cell Cycle and Proliferation. Antioxidants, 8(11), 552. https://doi.org/10.3390/antiox8110552