Differential Loss of OAS Genes Indicates Diversification of Antiviral Immunity in Mammals
Abstract
:1. Introduction
2. Materials and Methods
3. Results
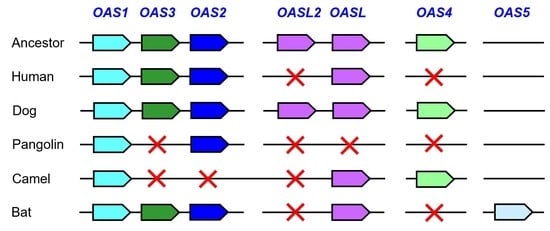
3.1. Comparative Genomics Reveals Loss of OAS Genes during the Evolution of Humans and Camelids
3.2. Pangolins have Lost Multiple OAS Genes
3.3. The Evolution of Bats Was Associated with the Diversification of OAS Paralogs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hornung, V.; Hartmann, R.; Ablasser, A.; Hopfner, K.P. OAS proteins and cGAS: Unifying concepts in sensing and responding to cytosolic nucleic acids. Nat. Rev. Immunol. 2014, 14, 521–528. [Google Scholar] [CrossRef]
- Hur, S. Double-stranded RNA sensors and modulators in innate immunity. Annu. Rev. Immunol. 2019, 37, 349–375. [Google Scholar] [CrossRef] [PubMed]
- Ablasser, A.; Hur, S. Regulation of cGAS- and RLR-mediated immunity to nucleic acids. Nat. Immunol. 2020, 21, 17–29. [Google Scholar] [CrossRef]
- Malathi, K.; Dong, B.; Gale MJr Silverman, R.H. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 2007, 448, 816–819. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Zhang, Y.; Ghosh, A.; Cuevas, R.A.; Forero, A.; Dhar, J.; Ibsen, M.S.; Schmid-Burgk, J.L.; Schmidt, T.; Ganapathiraju, M.K.; et al. Antiviral activity of human OASL protein is mediated by enhancing signaling of the RIG-I RNA sensor. Immunity 2014, 40, 936–948. [Google Scholar] [CrossRef]
- Rong, E.; Wang, X.; Chen, H.; Yang, C.; Hu, J.; Liu, W.; Wang, Z.; Chen, X.; Zheng, H.; Pu, J.; et al. Molecular mechanisms for the adaptive switching between the OAS/RNase L and OASL/RIG-I pathways in birds and mammals. Front. Immunol. 2018, 9, 1398. [Google Scholar] [CrossRef]
- Ghosh, A.; Shao, L.; Sampath, P.; Zhao, B.; Patel, N.V.; Zhu, J.; Behl, B.; Parise, R.A.; Beumer, J.H.; O’Sullivan, R.J.; et al. Oligoadenylate-synthetase-family protein OASL inhibits activity of the DNA sensor cGAS during DNA virus infection to limit interferon production. Immunity 2019, 50, 51–63.e5. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Wang, X.; Hu, J.; Song, L.; Rong, E.; Yang, C.; Chen, X.; Pu, J.; Sun, H.; Gao, C.; Burt, D.W.; et al. Functional divergence of oligoadenylate synthetase 1 (OAS1) proteins in tetrapods. Sci. China Life Sci. 2022, 65, 1395–1412. [Google Scholar] [CrossRef] [PubMed]
- Drappier, M.; Michiels, T. Inhibition of the OAS/RNase L pathway by viruses. Curr. Opin. Virol. 2015, 15, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Bignon, E.; Miclot, T.; Terenzi, A.; Barone, G.; Monari, A. Structure of the 5′ untranslated region in SARS-CoV-2 genome and its specific recognition by innate immune system via the human oligoadenylate synthase 1. Chem. Commun. 2022, 58, 2176–2179. [Google Scholar] [CrossRef] [PubMed]
- Hancks, D.C.; Hartley, M.K.; Hagan, C.; Clark, N.L.; Elde, N.C. Overlapping patterns of rapid evolution in the nucleic acid sensors cGAS and OAS1 suggest a common mechanism of pathogen antagonism and escape. PLoS Genet. 2015, 11, e1005203. [Google Scholar] [CrossRef] [PubMed]
- Mozzi, A.; Pontremoli, C.; Forni, D.; Clerici, M.; Pozzoli, U.; Bresolin, N.; Cagliani, R.; Sironi, M. OASes and STING: Adaptive evolution in concert. Genome Biol. Evol. 2015, 7, 1016–1032. [Google Scholar] [CrossRef]
- Lee, D.; Le Pen, J.; Yatim, A.; Dong, B.; Aquino, Y.; Ogishi, M.; Pescarmona, R.; Talouarn, E.; Rinchai, D.; Zhang, P.; et al. Inborn errors of OAS-RNase L in SARS-CoV-2-related multisystem inflammatory syndrome in children. Science 2023, 379, eabo3627. [Google Scholar] [CrossRef]
- Wickenhagen, A.; Sugrue, E.; Lytras, S.; Kuchi, S.; Noerenberg, M.; Turnbull, M.L.; Loney, C.; Herder, V.; Allan, J.; Jarmson, I.; et al. A prenylated dsRNA sensor protects against severe COVID-19. Science 2021, 374, eabj3624. [Google Scholar] [CrossRef] [PubMed]
- Carey, C.M.; Govande, A.A.; Cooper, J.M.; Hartley, M.K.; Kranzusch, P.J.; Elde, N.C. Recurrent loss-of-function mutations reveal costs to OAS1 antiviral activity in primates. Cell Host Microbe 2019, 25, 336–343. [Google Scholar] [CrossRef]
- Banday, A.R.; Stanifer, M.L.; Florez-Vargas, O.; Onabajo, O.O.; Papenberg, B.W.; Zahoor, M.A.; Mirabello, L.; Ring, T.J.; Lee, C.H.; Albert, P.S.; et al. Genetic regulation of OAS1 nonsense-mediated decay underlies association with COVID-19 hospitalization in patients of European and African ancestries. Nat. Genet. 2022, 54, 1103–1116. [Google Scholar] [CrossRef]
- Kjaer, K.H.; Poulsen, J.B.; Reintamm, T.; Saby, E.; Martensen, P.M.; Kelve, M.; Justesen, J. Evolution of the 2′-5′-oligoadenylate synthetase family in eukaryotes and bacteria. J. Mol. Evol. 2009, 69, 612–624. [Google Scholar] [CrossRef]
- Kumar, S.; Mitnik, C.; Valente, G.; Floyd-Smith, G. Expansion and molecular evolution of the interferon-induced 2′-5′ oligoadenylate synthetase gene family. Mol. Biol. Evol. 2000, 17, 738–750. [Google Scholar] [CrossRef]
- Perelygin, A.A.; Zharkikh, A.A.; Scherbik, S.V.; Brinton, M.A. The mammalian 2′-5′ oligoadenylate synthetase gene family: Evidence for concerted evolution of paralogous Oas1 genes in Rodentia and Artiodactyla. J. Mol. Evol. 2006, 63, 562–576. [Google Scholar] [CrossRef]
- Yao, Y.L.; Yu, D.; Xu, L.; Fan, Y.; Wu, Y.; Gu, T.; Chen, J.; Lv, L.B.; Yao, Y.G. Molecular characterization of the 2′,5′-oligoadenylate synthetase family in the Chinese tree shrew (Tupaia belangeri chinensis). Cytokine 2019, 114, 106–114. [Google Scholar] [CrossRef]
- Banerjee, A.; Baker, M.L.; Kulcsar, K.; Misra, V.; Plowright, R.; Mossman, K. Novel insights into immune systems of bats. Front. Immunol. 2020, 11, 26. [Google Scholar] [CrossRef] [PubMed]
- Fischer, H.; Tschachler, E.; Eckhart, L. Pangolins lack IFIH1/MDA5, a cytoplasmic RNA sensor that initiates innate immune defense upon coronavirus infection. Front. Immunol. 2020, 11, 939. [Google Scholar] [CrossRef]
- Fischer, H.; Tschachler, E.; Eckhart, L. Cytosolic DNA sensing through cGAS and STING is inactivated by gene mutations in pangolins. Apoptosis 2020, 25, 474–480. [Google Scholar] [CrossRef] [PubMed]
- Salova, M.; Sipos, W.; Tschachler, E.; Eckhart, L. NOD2 and reproduction-associated NOD-like receptors have been lost during the evolution of pangolins. Immunogenetics 2022, 74, 261–268. [Google Scholar] [CrossRef]
- Haley, P.J. From bats to pangolins: New insights into species differences in the structure and function of the immune system. Innate Immun. 2022, 28, 107–121. [Google Scholar] [CrossRef] [PubMed]
- Holmes, E.C.; Goldstein, S.A.; Rasmussen, A.L.; Robertson, D.L.; Crits-Christoph, A.; Wertheim, J.O.; Anthony, S.J.; Barclay, W.S.; Boni, M.F.; Doherty, P.C.; et al. The origins of SARS-CoV-2: A critical review. Cell 2021, 184, 4848–4856. [Google Scholar] [CrossRef]
- Lam, T.T.; Jia, N.; Zhang, Y.W.; Shum, M.H.; Jiang, J.F.; Zhu, H.C.; Tong, Y.G.; Shi, Y.X.; Ni, X.B.; Liao, Y.S.; et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 2020, 583, 282–285. [Google Scholar] [CrossRef] [PubMed]
- Eckhart, L.; Uthman, A.; Sipos, W.; Tschachler, E. Genome sequence comparison reveals independent inactivation of the caspase-15 gene in different evolutionary lineages of mammals. Mol. Biol. Evol. 2006, 23, 2081–2089. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Hedges, S.B.; Marin, J.; Suleski, M.; Paymer, M.; Kumar, S. Tree of life reveals clock-like speciation and diversification. Mol. Biol. Evol. 2015, 32, 835–845. [Google Scholar] [CrossRef]
- Azhar, E.I.; El-Kafrawy, S.A.; Farraj, S.A.; Hassan, A.M.; Al-Saeed, M.S.; Hashem, A.M.; Madani, T.A. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 2014, 370, 2499–2505. [Google Scholar] [CrossRef]
- Li, W.; Shi, Z.; Yu, M.; Ren, W.; Smith, C.; Epstein, J.H.; Wang, H.; Crameri, G.; Hu, Z.; Zhang, H.; et al. Bats are natural reservoirs of SARS-like coronaviruses. Science 2005, 310, 676–679. [Google Scholar] [CrossRef]
- Tian, J.; Sun, J.; Li, D.; Wang, N.; Wang, L.; Zhang, C.; Meng, X.; Ji, X.; Suchard, M.A.; Zhang, X.; et al. Emerging viruses: Cross-species transmission of coronaviruses, filoviruses, henipaviruses, and rotaviruses from bats. Cell Rep. 2022, 39, 110969. [Google Scholar] [CrossRef]
- Xie, J.; Li, Y.; Shen, X.; Goh, G.; Zhu, Y.; Cui, J.; Wang, L.F.; Shi, Z.L.; Zhou, P. Dampened STING-dependent interferon activation in bats. Cell Host Microbe 2018, 23, 297–301.e4. [Google Scholar] [CrossRef]
- Gonzalez, V.; Banerjee, A. Molecular, ecological, and behavioral drivers of the bat-virus relationship. iScience 2022, 25, 104779. [Google Scholar] [CrossRef]
- Irving, A.T.; Ahn, M.; Goh, G.; Anderson, D.E.; Wang, L.F. Lessons from the host defences of bats, a unique viral reservoir. Nature 2021, 589, 363–370. [Google Scholar] [CrossRef]
- Jacquet, S.; Culbertson, M.; Zhang, C.; El Filali, A.; De La Myre Mory, C.; Pons, J.B.; Filippi-Codaccioni, O.; Lauterbur, M.E.; Ngoubangoye, B.; Duhayer, J.; et al. Adaptive duplication and genetic diversification of protein kinase R contribute to the specificity of bat-virus interactions. Sci. Adv. 2022, 8, eadd7540. [Google Scholar] [CrossRef]
- Tsagkogeorga, G.; Parker, J.; Stupka, E.; Cotton, J.A.; Rossiter, S.J. Phylogenomic analyses elucidate the evolutionary relationships of bats. Curr. Biol. 2013, 23, 2262–2267. [Google Scholar] [CrossRef]
- Carelli, F.N.; Hayakawa, T.; Go, Y.; Imai, H.; Warnefors, M.; Kaessmann, H. The life history of retrocopies illuminates the evolution of new mammalian genes. Genome Res. 2016, 26, 301–314. [Google Scholar] [CrossRef]
- Rebouillat, D.; Hovanessian, A.G. The human 2′,5′-oligoadenylate synthetase family: Interferon-induced proteins with unique enzymatic properties. J. Interferon Cytokine Res. 1999, 19, 295–308. [Google Scholar] [CrossRef]
- Sadler, A.J.; Williams, B.R. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008, 8, 559–568. [Google Scholar] [CrossRef]
- Hu, J.; Wang, X.; Xing, Y.; Rong, E.; Ning, M.; Smith, J.; Huang, Y. Origin and development of oligoadenylate synthetase immune system. BMC Evol. Biol. 2018, 18, 201. [Google Scholar] [CrossRef] [PubMed]
- Mashimo, T.; Glaser, P.; Lucas, M.; Simon-Chazottes, D.; Ceccaldi, P.E.; Montagutelli, X.; Desprès, P.; Guénet, J.L. Structural and functional genomics and evolutionary relationships in the cluster of genes encoding murine 2′,5′-oligoadenylate synthetases. Genomics 2003, 82, 537–552. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Wu, X.; Shang, Y.; Wang, X.; Zhou, S.; Zhang, H. Adaptive evolution of the OAS gene family provides new insights into the antiviral ability of laurasiatherian mammals. Animals 2023, 13, 209. [Google Scholar] [CrossRef]
- Liu, H.L.; Yeh, I.J.; Phan, N.N.; Wu, Y.H.; Yen, M.C.; Hung, J.H.; Chiao, C.C.; Chen, C.F.; Sun, Z.; Jiang, J.Z.; et al. Gene signatures of SARS-CoV/SARS-CoV-2-infected ferret lungs in short- and long-term models. Infect Genet. Evol. 2020, 85, 104438. [Google Scholar] [CrossRef] [PubMed]
- Han, B.A.; Castellanos, A.A.; Schmidt, J.P.; Fischhoff, I.R.; Drake, J.M. The ecology of zoonotic parasites in the Carnivora. Trends Parasitol. 2021, 37, 1096–1110. [Google Scholar] [CrossRef] [PubMed]
- Sipos, W.; Lutonsky, C. Amendments suggested for zoo medical research strategies with focus on the D-A-CH region. Tierarztl. Prax. Ausg. G. Grosstiere Nutztiere 2021, 49, 256–260. [Google Scholar] [CrossRef]
- Mollentze, N.; Streicker, D.G. Viral zoonotic risk is homogenous among taxonomic orders of mammalian and avian reservoir hosts. Proc. Natl. Acad. Sci. USA. 2020, 117, 9423–9430. [Google Scholar] [CrossRef] [PubMed]
- MacNeill, A.L. Comparative pathology of zoonotic orthopoxviruses. Pathogens 2022, 11, 892. [Google Scholar] [CrossRef] [PubMed]
- Medina, R.A.; García-Sastre, A. Influenza A viruses: New research developments. Nat. Rev. Microbiol. 2011, 9, 590–603. [Google Scholar] [CrossRef] [PubMed]
- Short, E.C., Jr.; Fulton, R.W. Induction and measurement of 2′,5′-oligoadenylate synthetase in Madin-Darby bovine kidney cells and in cattle. J. Clin. Microbiol. 1987, 25, 1735–1740. [Google Scholar] [CrossRef]
- Azamor, T.; da Silva, A.M.V.; Melgaço, J.G.; Dos Santos, A.P.; Xavier-Carvalho, C.; Alvarado-Arnez, L.E.; Batista-Silva, L.R.; de Souza Matos, D.C.; Bayma, C.; Missailidis, S.; et al. Activation of an effective immune response after yellow fever vaccination is associated with the genetic background and early response of IFN-γ and CLEC5A. Viruses 2021, 13, 96. [Google Scholar] [CrossRef]
- Du, T.; Nan, Y.; Xiao, S.; Zhao, Q.; Zhou, E.M. Antiviral strategies against PRRSV infection. Trends Microbiol. 2017, 25, 968–979. [Google Scholar] [CrossRef] [PubMed]
- Indik, S.; Schmoll, F.; Sipos, W.; Klein, D. Genetic variability of PRRS virus in Austria: Consequences for molecular diagnostics and viral quantification. Vet. Microbiol. 2005, 107, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Sipos, W.; Duvigneau, C.; Pietschmann, P.; Höller, K.; Hartl, R.; Wahl, K.; Steinborn, R.; Gemeiner, M.; Willheim, M.; Schmoll, F. Parameters of humoral and cellular immunity following vaccination of pigs with a European modified-live strain of porcine reproductive and respiratory syndrome virus (PRRSV). Viral. Immunol. 2003, 16, 335–346. [Google Scholar] [CrossRef]
- Souvorov, A.; Kapustin, Y.; Kiryutin, B.; Chetvernin, V.; Tatusova, T.; Lipman, D. Gnomon—NCBI Eukaryotic Gene Prediction Tool. 2010. Available online: http://www.ncbi.nlm.nih.gov/RefSeq/Gnomon-description.pdf (accessed on 29 December 2022).
- Ehrlich, F.; Lachner, J.; Hermann, M.; Tschachler, E.; Eckhart, L. Convergent evolution of cysteine-rich keratins in hard skin appendages of terrestrial vertebrates. Mol. Biol. Evol. 2020, 37, 982–993. [Google Scholar] [CrossRef]
- Holthaus, K.B.; Lachner, J.; Ebner, B.; Tschachler, E.; Eckhart, L. Gene duplications and gene loss in the epidermal differentiation complex during the evolutionary land-to-water transition of cetaceans. Sci. Rep. 2021, 11, 12334. [Google Scholar] [CrossRef]
- McDougal, M.B.; Boys, I.N.; De La Cruz-Rivera, P.; Schoggins, J.W. Evolution of the interferon response: Lessons from ISGs of diverse mammals. Curr. Opin. Virol. 2022, 53, 101202. [Google Scholar] [CrossRef] [PubMed]
- Glidden, C.K.; Nova, N.; Kain, M.P.; Lagerstrom, K.M.; Skinner, E.B.; Mandle, L.; Sokolow, S.H.; Plowright, R.K.; Dirzo, R.; De Leo, G.A.; et al. Human-mediated impacts on biodiversity and the consequences for zoonotic disease spillover. Curr. Biol. 2021, 31, R1342–R1361. [Google Scholar] [CrossRef]






| Species | Binomial Name | OAS1 | OAS2 | OAS3 | OAS4 | OAS5 | OASL | OASL2 |
|---|---|---|---|---|---|---|---|---|
| Human | Homo sapiens | + | + | + | – | – | + | – |
| Bactrian camel | Camelus bactrianus | + | – | – | + | – | + | – |
| Arabian camel | Camelus dromedarius | + | – | – | + | – | + | – |
| Alpaca | Vicugna pacos | + | – | – | + | – | + | – |
| Dog | Canis familiaris | + | + | + | + | – | + | + |
| Malayan pangolin | Manis javanica | + * | + | – | – | – | – | – |
| Chinese pangolin | Manis pentadactyla | + | + | – | – | – | – | – |
| Chinese rufous horseshoe bat | Rhinolophus sinicus | + | + | + | – | – | + | + |
| Pallas’s mastiff bat | Molossus molossus | + * | + | + | – | + | + | – |
| Greater mouse-eared bat | Myotis myotis | + | + | + | – | + | + | – |
| Jamaican fruit bat | Artibeus jamaicensis | + | + * | + | – | + | + | – |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eckhart, L.; Sipos, W. Differential Loss of OAS Genes Indicates Diversification of Antiviral Immunity in Mammals. Vaccines 2023, 11, 419. https://doi.org/10.3390/vaccines11020419
Eckhart L, Sipos W. Differential Loss of OAS Genes Indicates Diversification of Antiviral Immunity in Mammals. Vaccines. 2023; 11(2):419. https://doi.org/10.3390/vaccines11020419
Chicago/Turabian StyleEckhart, Leopold, and Wolfgang Sipos. 2023. "Differential Loss of OAS Genes Indicates Diversification of Antiviral Immunity in Mammals" Vaccines 11, no. 2: 419. https://doi.org/10.3390/vaccines11020419
APA StyleEckhart, L., & Sipos, W. (2023). Differential Loss of OAS Genes Indicates Diversification of Antiviral Immunity in Mammals. Vaccines, 11(2), 419. https://doi.org/10.3390/vaccines11020419