A Novel Combined Scientific and Artistic Approach for the Advanced Characterization of Interactomes: The Akirin/Subolesin Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. Approaching Collaboration between Visual and Musical Artists and Scientists
2.2. Yeast Two-Hybrid Y2H Screening for the Identification of Human AKR2 Interacting Proteins
2.3. Interaction Confidence Scoring
- A: Very high confidence in the interaction.
- B: High confidence in the interaction.
- C: Good confidence in the interaction.
- D: Moderate confidence in the interaction. This category is the most difficult to interpret because it mixes two classes of interactions: (i) false-positive interactions and (ii) interactions hardly detectable by the Y2H technique (e.g., low representation of the mRNA in the library, prey folding, prey toxicity in yeast).
- E: Interactions involving highly connected (or relatively highly connected) prey domains, warning of non-specific interaction. The total number of screens performed on each organism is taken into account to set this connectivity threshold to 20 interactions to different bait proteins in the entire human database. They can be classified in different categories: (i) prey proteins that are known to be highly connected due to their biological function and (ii) proteins with a prey interacting domain that contains a known protein interaction motif or a biochemically promiscuous motif.
- F: Experimentally proven technical artifacts.
- N/A: The PBS is a score that is automatically computed through algorithms and cannot be attributed for the following reasons: (i) all the fragments of the same reference coding sequence (CDS) are antisense, (ii) the 5p sequence is missing, (iii) all the fragments of the same reference CDS are either all out of frame (OOF1 or OOF2), and (iv) all the fragments of the same reference CDS lie in the 5′ or 3′ untranslated region (UTR).
2.4. Annotation of Identified AKR2-Interacting Proteins
2.5. Corroboration of AKR2–Protein Interactions by Protein Pull-Down
2.6. In Vitro Characterization of AKR–AKR Protein Interactions
2.7. Prediction Models of Protein–Protein Binding Sites and Structure
2.8. Musical Scores and Ensembles
2.9. NF-κB Reporter Assay
2.10. Gene Knockdown by RNA Interference (RNAi)
2.11. Treatment of Human Placenta Cells with Lipopolysaccharides (LPS)
2.12. Analysis of Gene Expression by qRT-PCR
2.13. Protein Transfection
2.14. Prediction and Characterization of AKR/SUB Protective Epitopes
3. Results
3.1. Artist New Perspectives AKR Dimerization and Multiple Simultaneous AKR2–Protein Interactions with Functional Implications
3.2. The Sound of the AKR/SUB Coding Sequence Supported Evolutionary Conservation and Functional Protein Interactions
3.3. AKR2–Protein Interactions Positively and Negatively Regulated the NF-κB Signaling Pathway
3.4. LPS Did Not Activate the NF-κB Pathway Via AKR2–Protein Interactions in Human Placenta Cells
3.5. The Characterization of the AKR/SUB Interactome Had Implications for Quantum Vaccinomics
4. Discussions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jeffries, S. When two tribes meet: Collaborations between artists and scientists. The Guardian. 21 August 2011. Available online: https://www.theguardian.com/artanddesign/2011/aug/21/collaborations-between-artists-and-scientists (accessed on 1 August 2019).
- Tayag, Y.; Wells, B. Art and evolution: A work in progress. Scientific American. 6 May 2014. Available online: https://blogs.scientificamerican.com/guest-blog/art-and-evolution-a-work-in-progress/ (accessed on 1 August 2019).
- Schulkin, J.; Raglan, G.B. The evolution of music and human social capability. Front. Neurosci. 2014, 8, 292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de la Fuente, J.; Estrada-Peña, A.; Cabezas-Cruz, A.; Brey, R. Flying ticks: Anciently evolved associations that constitute a risk of infectious disease spread. Parasit. Vectors 2015, 8, 538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veis, N. Four takes in the evolution of art. Nature 2017, 543, 490. [Google Scholar] [CrossRef] [Green Version]
- Stevens, C.; O’Connor, G. When artists get involved in research, science benefits. The Conversation. 16 August 2017. Available online: http://theconversation.com/when-artists-get-involved-in-research-science-benefits-82147 (accessed on 1 August 2019).
- de la Fuente, J. Anaplasmosis: What we can learn from Lam’s surrealistic animalarium. Hektoen International Hektorama-Infectious Diseases-Summer. 2018. Available online: http://hekint.org/2018/08/23/anasplasmosis-what-we-can-learn-from-lams-surrealistic-animalarium/ (accessed on 1 August 2019).
- Riego, E.; Silva, A.; de la Fuente, J. The sound of the DNA language. Biol. Res. 1995, 28, 197–204. [Google Scholar] [PubMed]
- Eldred, S.M. Art–science collaborations: Change of perspective. Nature 2016, 537, 125–126. [Google Scholar] [CrossRef] [Green Version]
- Shelton, J. A visual artist compares the way scientists and artists see a world of discovery. YaleNews. 3 April 2017. Available online: https://news.yale.edu/2017/04/03/visual-artist-compares-way-scientists-and-artists-see-world-discovery (accessed on 1 August 2019).
- Maeda, J. Artists and scientists: More alike than different. Scientific American. 11 July 2013. Available online: https://blogs.scientificamerican.com/guest-blog/artists-and-scientists-more-alike-than-different/ (accessed on 1 August 2019).
- Peña-Rangel, M.T.; Rodriguez, I.; Riesgo-Escovar, J.R. A misexpression study examining dorsal thorax formation in Drosophila melanogaster. Genetics 2002, 160, 1035–1050. [Google Scholar]
- Almazán, C.; Kocan, K.M.; Bergman, D.K.; Garcia-Garcia, J.C.; Blouin, E.F.; de la Fuente, J. Identification of protective antigens for the control of Ixodes scapularis infestations using cDNA expression library immunization. Vaccine 2003, 21, 1492–1501. [Google Scholar] [CrossRef]
- DasGupta, R.; Kaykas, A.; Moon, R.T.; Perrimon, N. Functional genomic analysis of the Wnt-wingless signaling pathway. Science 2005, 308, 826–833. [Google Scholar] [CrossRef] [Green Version]
- Goto, A.; Matsushita, K.; Gesellchen, V.; El Chamy, L.; Kuttenkeuler, D.; Takeuchi, O.; Hoffmann, J.A.; Akira, S.; Boutros, M.; Reichhart, J.M. Akirins are highly conserved nuclear proteins required for NF-kappaB-dependent gene expression in Drosophila and mice. Nat. Immunol. 2008, 9, 97–104. [Google Scholar] [CrossRef] [Green Version]
- Macqueen, D.J.; Johnston, I.A. Evolution of the multifaceted eukaryotic akirin gene family. BMC Evol. Biol. 2009, 9, 34. [Google Scholar] [CrossRef] [Green Version]
- Artigas-Jerónimo, S.; Villar, M.; Cabezas-Cruz, A.; Valdés, J.J.; Estrada-Peña, A.; Alberdi, P.; de la Fuente, J. Functional evolution of Subolesin/Akirin. Front. Physiol. 2018, 9, 1612. [Google Scholar] [CrossRef] [PubMed]
- Polanowska, J.; Chen, J.X.; Soulé, J.; Omi, S.; Belougne, J.; Taffoni, C.; Pujol, N.; Selbach, M.; Zugasti, O.; Ewbank, J.J. Evolutionary plasticity in the innate immune function of Akirin. PLoS Genet. 2018, 14, e1007494. [Google Scholar] [CrossRef] [Green Version]
- Rual, J.F.; Venkatesan, K.; Hao, T.; Hirozane-Kishikawa, T.; Dricot, A.; Li, N.; Berriz, G.F.; Gibbons, F.D.; Dreze, M.; Ayivi-Guedehoussou, N.; et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature 2005, 437, 1173–1178. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, K.; Srikantan, S.; Yang, X.; Lal, A.; Kim, H.H.; Kuwano, Y.; Galban, S.; Becker, K.G.; Kamara, D.; de Cabo, R.; et al. Ubiquitin-mediated proteolysis of HuR by heat shock. EMBO J. 2009, 28, 1271–1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armour, S.M.; Bennett, E.J.; Braun, C.R.; Zhang, X.Y.; McMahon, S.B.; Gygi, S.P.; Harper, J.W.; Sinclair, D.A. A high-confidence interaction map identifies SIRT1 as a mediator of acetylation of USP22 and the SAGA coactivator complex. Mol. Cell. Biol. 2013, 33, 1487–1502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hein, M.Y.; Hubner, N.C.; Poser, I.; Cox, J.; Nagaraj, N.; Toyoda, Y.; Gak, I.A.; Weisswange, I.; Mansfeld, J.; Buchholz, F.; et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 2015, 163, 712–723. [Google Scholar] [CrossRef] [Green Version]
- Huttlin, E.L.; Bruckner, R.J.; Paulo, J.A.; Cannon, J.R.; Ting, L.; Baltier, K.; Colby, G.; Gebreab, F.; Gygi, M.P.; Parzen, H.; et al. Harper, architecture of the human interactome defines protein communities and disease networks. Nature 2017, 545, 505–509. [Google Scholar] [CrossRef]
- de la Fuente, J.; Maritz-Olivier, C.; Naranjo, V.; Ayoubi, P.; Nijhof, A.M.; Almazán, C.; Canales, M.; Pérez de la Lastra, J.M.; Galindo, R.C.; Blouin, E.F.; et al. Evidence of the role of tick subolesin in gene expression. BMC Genom. 2008, 9, 372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowak, S.J.; Baylies, M.K. Akirin: A context-dependent link between transcription and chromatin remodeling. Bioarchitecture 2012, 2, 209–213. [Google Scholar] [CrossRef]
- Goto, A.; Fukuyama, H.; Imler, J.L.; Hoffmann, J.A. The chromatin regulator DMAP1 modulates activity of the nuclear factor B (NF-B) transcription factor Relish in the Drosophila innate immune response. J. Biol. Chem. 2014, 289, 20470–20476. [Google Scholar] [CrossRef] [Green Version]
- Bonnay, F.; Nguyen, X.H.; Cohen-Berros, E.; Troxler, L.; Batsche, E.; Camonis, J.; Takeuchi, O.; Reichhart, J.M.; Matt, N. Akirin specifies NF-κB selectivity of Drosophila innate immune response via chromatin remodeling. EMBO J. 2014, 33, 2349–2362. [Google Scholar] [CrossRef] [PubMed]
- Shaw, D.K.; Wang, X.; Brown, L.J.; Chávez, A.S.; Reif, K.E.; Smith, A.A.; Scott, A.J.; McClure, E.E.; Boradia, V.M.; Hammond, H.L.; et al. Infection-derived lipids elicit an immune deficiency circuit in arthropods. Nat. Commun. 2017, 8, 14401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de la Fuente, J.; Kopáček, P.; Lew-Tabor, A.; Maritz-Olivier, C. Strategies for new and improved vaccines against ticks and tick-borne diseases. Parasite Immunol. 2016, 38, 754–769. [Google Scholar] [CrossRef] [PubMed]
- Camnitzer, L. New Art of Cuba; University of Texas Press: Austin, TX, USA, 1994. [Google Scholar]
- Cordero, R. Raúl Cordero; Turner: Madrid, Spain, 2010. [Google Scholar]
- Formstecher, E.; Aresta, S.; Collura, V.; Hamburger, A.; Meil, A.; Trehin, A.; Reverdy, C.; Betin, V.; Maire, S.; Brun, C.; et al. Protein interaction mapping: A Drosophila case study. Genome Res. 2005, 15, 376–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rain, J.C.; Selig, L.; De Reuse, H.; Battaglia, V.; Reverdy, C.; Simon, S.; Lenzen, G.; Petel, F.; Wojcik, J.; Schächter, V.; et al. The protein-protein interaction map of Helicobacter pylori. Nature 2011, 409, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Cid, J.A.; Pérez de la Lastra, J.M.; Villar, M.; Jiménez, M.; Pinal, R.; Estrada-Peña, A.; Molina, R.; Lucientes, J.; Gortázar, C.; de la Fuente, J. SUB/AKR Vaccine Study Group. Control of multiple arthropod vector infestations with subolesin/akirin vaccines. Vaccine 2013, 31, 1187–1196. [Google Scholar] [CrossRef]
- Merino, O.; Antunes, S.; Mosqueda, J.; Moreno-Cid, J.A.; Pérez de la Lastra, J.M.; Rosario-Cruz, R.; Rodríguez, S.; Domingos, A.; de la Fuente, J. Vaccination with proteins involved in tick-pathogen interactions reduces vector infestations and pathogen infection. Vaccine 2013, 31, 5889–5896. [Google Scholar] [CrossRef]
- Garcia-Garcia, J.; Valls-Comamala, V.; Guney, E.; Andreu, D.; Muñoz, F.J.; Fernandez-Fuentes, N.; Oliva, B. iFrag: A protein-protein interface prediction server based on sequence fragments. J. Mol. Biol. 2017, 429, 382–389. [Google Scholar] [CrossRef]
- Ayllón, N.; Villar, M.; Busby, A.T.; Kocan, K.M.; Blouin, E.F.; Bonzón-Kulichenko, E.; Galindo, R.C.; Mangold, A.J.; Alberdi, P.; Pérez de la Lastra, J.M.; et al. Anaplasma phagocytophilum inhibits apoptosis and promotes cytoskeleton rearrangement for infection of tick cells. Infect. Immun. 2013, 81, 2415–2425. [Google Scholar] [CrossRef] [Green Version]
- Prudencio, C.R.; Pérez de la Lastra, J.M.; Canales, M.; Villar, M.; de la Fuente, J. Mapping protective epitopes in the tick and mosquito subolesin ortholog proteins. Vaccine 2010, 28, 5398–5406. [Google Scholar] [CrossRef]
- Ward, J.J.; Sodhi, J.S.; McGuffin, L.J.; Buxton, B.F.; Jones, D.T. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 2004, 337, 635–645. [Google Scholar] [CrossRef]
- Kim, D.E.; Chivian, D.; Baker, D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004, 32, W526–W531. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Zhang, Y. LOMETS: A local meta-threading-server for protein structure prediction. Nucleic Acids Res. 2007, 35, 3375–3382. [Google Scholar] [CrossRef] [Green Version]
- Benkert, P.; Kunzli, M.; Schwede, T. QMEAN server for protein model quality estimation. Nucleic Acids Res. 2009, 37, W510–W514. [Google Scholar] [CrossRef] [Green Version]
- Wingelhofer, B.; Neubauer, H.A.; Valent, P.; Han, X.; Constantinescu, S.N.; Gunning, P.T.; Müller, M.; Moriggl, R. Implications of STAT3 and STAT5 signaling on gene regulation and chromatin remodeling in hematopoietic cancer. Leukemia 2018, 32, 1713–1726. [Google Scholar] [CrossRef] [Green Version]
- van Essen, D.; Engist, B.; Natoli, G.; Saccani, S. Two modes of transcriptional activation at native promoters by NF-kappaB p65. PLoS Biol. 2009, 7, e73. [Google Scholar] [CrossRef] [Green Version]
- Tieri, P.; Termanini, A.; Bellavista, E.; Salvioli, S.; Capri, M.; Franceschi, C. Charting the NF-κB pathway interactome map. PLoS ONE 2012, 7, e32678. [Google Scholar] [CrossRef] [Green Version]
- Joly, S.; Rhea, L.; Volk, P.; Moreland, J.G.; Dunnwald, M. Interferon regulatory factor 6 has a protective role in the host response to endotoxic shock. PLoS ONE 2016, 11, e0152385. [Google Scholar] [CrossRef] [Green Version]
- Pranski, E.L.; Dalal, N.V.; Herskowitz, J.H.; Orr, A.L.; Roesch, L.A.; Fritz, J.J.; Heilman, C.; Lah, J.J.; Levey, A.I.; Betarbet, R.S. Neuronal RING finger protein 11 (RNF11) regulates canonical NF-κB signaling. J. Neuroinflamm. 2012, 9, 67. [Google Scholar] [CrossRef] [Green Version]
- Frasor, J.; El-Shennawy, L.; Stender, J.D.; Kastrati, I. NFκB affects estrogen receptor expression and activity in breast cancer through multiple mechanisms. Mol. Cell. Endocrinol. 2015, 418 Pt 3, 235–239. [Google Scholar] [CrossRef] [Green Version]
- Du, Q.; Geller, D.A. Cross-regulation between Wnt and NF-κB signaling pathways. For Immunopathol. Dis. Therap. 2010, 1, 155–181. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.; Baltimore, D. Circuitry of nuclear factor kappaB signaling. Immunol. Rev. 2006, 210, 171–186. [Google Scholar] [CrossRef] [PubMed]
- de la Garza, G.; Schleiffarth, J.R.; Dunnwald, M.; Mankad, A.; Weirather, J.L.; Bonde, G.; Butcher, S.; Mansour, T.A.; Kousa, Y.A.; Fukazawa, C.F.; et al. Interferon regulatory factor 6 promotes differentiation of the periderm by activating expression of Grainyhead-like 3. J. Investig. Dermatol. 2013, 133, 68–77. [Google Scholar] [CrossRef] [Green Version]
- Kowalec, K.; Wright, G.E.B.; Drögemöller, B.I.; Aminkeng, F.; Bhavsar, A.P.; Kingwell, E.; Yoshida, E.M.; Traboulsee, A.; Marrie, R.A.; Kremenchutzky, M.; et al. Common variation near IRF6 is associated with IFN-β-induced liver injury in multiple sclerosis. Nat. Genet. 2018, 50, 1081–1085. [Google Scholar] [CrossRef]
- Lenardo, M.J.; Fan, C.M.; Maniatis, T.; Baltimore, D. The involvement of NF-κB in β-interferon gene regulation reveals its role as widely inducible mediator of signal transduction. Cell 1989, 57, 287–294. [Google Scholar] [CrossRef]
- MacDonald, N.J.; Kuhl, D.; Maguire, D.; Näf, D.; Gallant, P.; Goswamy, A.; Hug, H.; Büeler, H.; Chaturvedi, M.; de la Fuente, J.; et al. Different pathways mediate virus inducibility of the human IFN-alpha1 and IFN-beta genes. Cell 1990, 60, 767–779. [Google Scholar] [CrossRef]
- Hiscott, J.; Alper, D.; Cohen, L.; Leblanc, J.F.; Sportza, L.; Wong, A.; Xanthoudakis, S. Induction of human interferon gene expression is associated with a nuclear factor that interacts with the NF-kappa B site of the human immunodeficiency virus enhancer. J. Virol. 1989, 63, 2557–2566. [Google Scholar] [CrossRef] [Green Version]
- Sharif, O.; Bolshakov, V.N.; Raines, S.; Newham, P.; Perkins, N.D. Transcriptional profiling of the LPS induced NF-kB response in macrophages. BMC Immunol. 2007, 8, 1471–2172. [Google Scholar] [CrossRef] [Green Version]
- Sheikh, F.; Dickensheets, H.; Gamero, A.M.; Vogel, S.N.; Donnelly, R.P. An essential role for IFN-β in the induction of IFN-stimulated gene expression by LPS in macrophages. J. Leukoc. Biol. 2014, 96, 591–600. [Google Scholar] [CrossRef]
- Ashdown, H.; Dumont, Y.; Ng, M.; Poole, S.; Boksa, P.; Luheshi, G.N. The role of cytokines in mediating effects of prenatal infection on the fetus: Implications for schizophrenia. Mol. Psychiatry 2006, 11, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Sakai, J.; Cammarota, E.; Wright, J.A.; Cicuta, P.; Gottschalk, R.A.; Li, N.; Fraser, I.D.C.; Bryant, C.E. Lipopolysaccharide-induced NF-κB nuclear translocation is primarily dependent on MyD88, but TNFα expression requires TRIF and MyD88. Sci Rep. 2017, 7, 1428. [Google Scholar] [CrossRef]
- Contreras, M.; de la Fuente, J. Control of Ixodes ricinus and Dermacentor reticulatus tick infestations in rabbits vaccinated with the Q38 Subolesin/Akirin chimera. Vaccine 2016, 34, 3010–3013. [Google Scholar] [CrossRef]
- Ailenberg, M.; Rotstein, O. An improved Huffman coding method for archiving text, images, and music characters in DNA. Biotechniques 2009, 47, 747–754. [Google Scholar] [CrossRef]
- Temple, M.D. An auditory display tool for DNA sequence analysis. BMC Bioinform. 2017, 18, 221. [Google Scholar] [CrossRef]
- Mannone, M. Knots, music and DNA. J. Creat. Music Syst. 2018, 2, 32. [Google Scholar] [CrossRef] [Green Version]
- de la Fuente, J.; Estrada-Peña, A. Why new vaccines for the control of ectoparasite vectors have not been registered and commercialized? Vaccines 2019, 7, 75. [Google Scholar] [CrossRef] [Green Version]
- de la Fuente, J.; Moreno-Cid, J.A.; Canales, M.; Villar, M.; Pérez de la Lastra, J.M.; Kocan, K.M.; Galindo, R.C.; Almazán, C.; Blouin, E.F. Targeting arthropod subolesin/akirin for the development of a universal vaccine for control of vector infestations and pathogen transmission. Vet. Parasitol. 2011, 181, 17–22. [Google Scholar] [CrossRef]
- de la Fuente, J.; Moreno-Cid, J.A.; Galindo, R.C.; Almazán, C.; Kocan, K.M.; Merino, O.; Pérez de la Lastra, J.M.; Estrada-Peña, A.; Blouin, E.F. Subolesin/Akirin vaccines for the control of arthropod vectors and vector-borne pathogens. Transbound. Emerg. Dis. 2013, 60 (Suppl. 2), 172–178. [Google Scholar] [CrossRef]
- de la Fuente, J.; Merino, O. Vaccinomics, the new road to tick vaccines. Vaccine 2013, 31, 5923–5929. [Google Scholar] [CrossRef]
- Contreras, M.; Villar, M.; de la Fuente, J. A vaccinomics approach to the identification of tick protective antigens for the control of Ixodes ricinus and Dermacentor reticulatus infestations in companion animals. Front. Physiol. 2019, 10, 977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Artigas-Jerónimo, S.; Estrada-Peña, A.; Cabezas-Cruz, A.; Alberdi, P.; Villar, M.; de la Fuente, J. Modeling modulation of the tick regulome in response to Anaplasma phagocytophilum for the identification of new control targets. Front. Physiol. 2019, 10, 462. [Google Scholar] [CrossRef] [PubMed]
- de la Fuente, J.; Villar, M.; Estrada-Peña, A.; Olivas, J.A. High throughput discovery and characterization of tick and pathogen vaccine protective antigens using vaccinomics with intelligent Big Data analytic techniques. Expert Rev. Vaccines 2018, 17, 569–576. [Google Scholar] [CrossRef]









| Name | Description | Protein/Gene ID | Interaction Score (PBS) |
|---|---|---|---|
| AKR1 § | Akirin1 | Q9H9L7 | A |
| ACTR10 | Actin related protein 10 | Q9NZ32 | A |
| RNF10 § | RING finger protein 10 | Q8N5U6 | A |
| SF3A1 | Splicing factor 3a subunit 1 | Q15459 | A |
| THRAP5 § | Mediator of RNA polymerase II transcription subunit 16 | Q9Y2X0 | A |
| AKR2 § | Akirin2 | Q53H80 | B |
| ESRRG § | Estrogen related receptor gamma | P62508 | B |
| PITPNA | Phosphatidylinositol transfer protein alpha isoform | Q00169 | B |
| ATF4 | Cyclic AMP-dependent transcription factor ATF-4 | P18848 | D |
| C12orf10 | Chromosome 12 open reading frame 10 | Q86UA3 | D |
| CCNE1 | G1/S-specific cyclin-E1 | P24864 | D |
| CDC23 | Cell division cycle protein 23 homolog | Q9UJX2 | D |
| CLIC3 | Chloride intracellular channel protein 3 | O95833 | D |
| CLTCL1 | Clathrin heavy chain 2 | P53675 | D |
| CNOT3 | CCR4-NOT transcription complex subunit 3 | O75175 | D |
| EXPH5 | Exophilin 5 | Q149M6 | D |
| EZR | Ezrin | P15311 | D |
| FNTA | Protein farnesyltransferase/geranylgeranyltransferase type-1 subunit alpha | P49354 | D |
| HEG1 | Protein HEG homolog 1 | Q9ULI3 | D |
| HNRPF | Heterogeneous nuclear ribonucleoprotein F | P52597 | D |
| IFT57 | Intraflagellar transport protein 57 homolog | Q9NWB7 | D |
| INTS2 | Integrator complex subunit 2 | Q9H0H0 | D |
| IRF6 § | Interferon regulatory factor 6 | O14896 | D |
| KLHL5 | Kelch-like 5 isoform 2 variant | Q59HD9 | D |
| LIMS1 | LIM and senescent cell antigen-like-containing domain protein 1 | P48059 | D |
| LNX1 | E3 ubiquitin-protein ligase LNX | Q8TBB1 | D |
| PKM | Pyruvate kinase PKM | P14618 | D |
| SCFD1 | Sec1 family domain-containing protein 1 | Q8WVM8 | D |
| SERPINE2 | Glia-derived nexin | P07093 | D |
| SF3B1 | Splicing factor 3B subunit 1 | O75533 | D |
| SPG21 | Maspardin | Q9NZD8 | D |
| SPTBN1 var 1 | Spectrin beta chain, non-erythrocytic 1 | Q01082 | D |
| TOPBP1 | DNA topoisomerase 2-binding protein 1 | Q92547 | D |
| URB2 | Unhealthy ribosome biogenesis protein 2 homolog | Q14146 | D |
| USP7 | Ubiquitin carboxyl-terminal hydrolase 7 | Q93009 | D |
| VPS39 | Vam6/Vps39-like protein | Q96JC1 | D |
| WNT2 § | Protein Wnt-2 | P09544 | D |
| ZNF862 | Zinc finger protein 862 | O60290 | D |
| GAREM1 * | GRB2-associated and regulator of MAPK protein1 | Q9H706 | D |
| Mitochondrion * | Mitochondrion, complete genome | MF992925 | D |
| RBPMS * | RNA binding protein, mRNA processing factor (RBPMS), on chromosome 8 | NG_029534.1 | D |
| BRD7 * | Bromodomain containing 7 (BRD7), on chromosome 16 | NG_023418.1 | D |
| LSS * | Lanosterol synthase (LSS), on chromosome 21 | NG_011510.1 | D |
| DRC3 * | DRC3 gene, complete cds | AF282168.1 | D |
| AF-6 * | AF-6, complete cds | AB011399.1 | D |
| DAPK1 * | Death-associated protein kinase 1 (DAPK1) gene, complete cds | DQ436495.1 | D |
| EDH17B2 * | 17-beta-hydroxysteroid dehydrogenase (EDH17B2) gene, complete cds | U34879.1 | D |
| Artist Contribution | Scientist Perspective |
|---|---|
| Methodological Approach | |
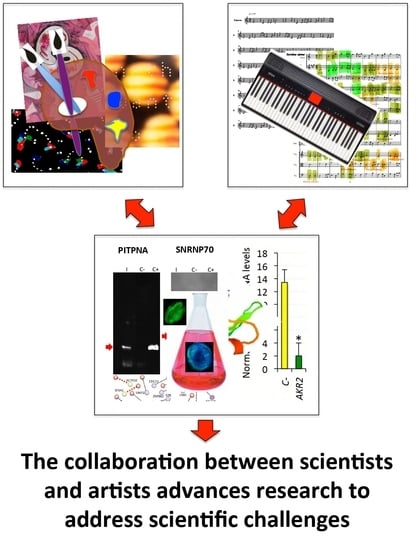
| Three visual artists and a musician were invited to read a simplified version of our recent review on AKR/SUB functional evolution [17] and were challenged with the conserved function of these proteins in the regulation of different biological processes throughout the metazoan. | In response to this challenge, the artists contributed the pieces, musical scores, and interpretations shown in the paper in order to provide their view on this matter. Artists’ contributions served to inspire scientists to discuss and find new perspectives on unexplored characteristics of these proteins with putative functional implications. |
| Piece in Figure 1A | |
| The artist represents the origins of life, with multiple geometric images that interact to illustrate in different species the conserved function of these proteins in biological processes represented by the sea, paper boat, Picasso’s dove, and a fetus growing in a mother’s womb while opening its eyes to the world. | From a scientist’s perspective, a new and unexplored facet of possible functional relevance of AKR dimerization was proposed in this piece. |
| Piece in Figure 2A | |
| The artist describes the constant movement of these proteins that translate into the visual rhythm of repetitive interconnected interactions of forms and colors. | From a scientist’s perspective, the possibility that AKR physically interacts with different proteins simultaneously to regulate various biological processes defined by cell-specific AKR–protein interactions was proposed in this piece. |
| Piece in Figure 6A | |
| The artist proposed that what you are looking for is also looking for you, highlighting that nothing wants to tell you something and it is only a matter of finding how to perceive the message. | From a scientist’s perspective, this message challenges the view that AKR–protein interactions occur randomly and suggests that these interactions are functionally relevant in the regulation of different biological processes as occurs with other regulatory factors. |
| Musical Scores in Figure 5 and Figures S2–S4 | |
| A revised algorithm using musical ensembles was applied to translate the AKR and interacting protein sequences into music. | The results supported that AKR in different species are evolutionarily related and structurally conserved, and confirmed AKR2–protein interactions identified in this study. Therefore, this tool may be used for evolutionary studies and the prediction of protein–protein interactions. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Artigas-Jerónimo, S.; Pastor Comín, J.J.; Villar, M.; Contreras, M.; Alberdi, P.; León Viera, I.; Soto, L.; Cordero, R.; Valdés, J.J.; Cabezas-Cruz, A.; et al. A Novel Combined Scientific and Artistic Approach for the Advanced Characterization of Interactomes: The Akirin/Subolesin Model. Vaccines 2020, 8, 77. https://doi.org/10.3390/vaccines8010077
Artigas-Jerónimo S, Pastor Comín JJ, Villar M, Contreras M, Alberdi P, León Viera I, Soto L, Cordero R, Valdés JJ, Cabezas-Cruz A, et al. A Novel Combined Scientific and Artistic Approach for the Advanced Characterization of Interactomes: The Akirin/Subolesin Model. Vaccines. 2020; 8(1):77. https://doi.org/10.3390/vaccines8010077
Chicago/Turabian StyleArtigas-Jerónimo, Sara, Juan J. Pastor Comín, Margarita Villar, Marinela Contreras, Pilar Alberdi, Israel León Viera, Leandro Soto, Raúl Cordero, James J. Valdés, Alejandro Cabezas-Cruz, and et al. 2020. "A Novel Combined Scientific and Artistic Approach for the Advanced Characterization of Interactomes: The Akirin/Subolesin Model" Vaccines 8, no. 1: 77. https://doi.org/10.3390/vaccines8010077
APA StyleArtigas-Jerónimo, S., Pastor Comín, J. J., Villar, M., Contreras, M., Alberdi, P., León Viera, I., Soto, L., Cordero, R., Valdés, J. J., Cabezas-Cruz, A., Estrada-Peña, A., & de la Fuente, J. (2020). A Novel Combined Scientific and Artistic Approach for the Advanced Characterization of Interactomes: The Akirin/Subolesin Model. Vaccines, 8(1), 77. https://doi.org/10.3390/vaccines8010077