Deciphering the Mechanisms of Improved Immunogenicity of Hypochlorous Acid-Treated Antigens in Anti-Cancer Dendritic Cell-Based Vaccines
Abstract
:1. Introduction
2. Materials and Methods
2.1. Whole Tumor Lysate (WTL) Preparation
2.2. DC Preparation and WTL Loading
2.3. HLA Typing
2.4. Sample Preparation for Proteomics Analysis
2.5. Immunoaffinity Purification of HLA Peptides
2.6. LC-MS/MS Analyses
2.7. Peptide and Protein Identification and Bioinformatic Analyses
2.8. T Cell Phenotypic and Functional Analysis
2.9. Statistics
2.10. Data Availability
3. Results
3.1. HOCl Incubation Induces Extensive Amino Acid Oxidation in Tumor Cells
3.2. HOCl Induces Specific Patterns of Protein Expression and Oxidation in Tumor Cells
3.3. HOCl Antigen Treatment Does not Significantly Affect DC Phenotype
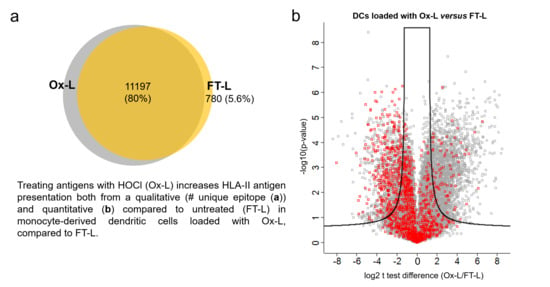
3.4. HOCl Antigen Treatment Significantly Improves MHC-II Antigen Presentation in Mo-DCs
3.5. Solvent Exposed Regions in Source Proteins upon HOCl Treatment Contribute to the Mo-DCs HLA-II Ligandome
3.6. Vaccination with Mo-DCs Antigen-Stimulated with Autologous Ox-L Activates CD4+ T Cell Responses in Ovarian Cancer Patients, in Adjuvant Settings
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Banchereau, J.; Briere, F.; Caux, C.; Davoust, J.; Lebecque, S.; Liu, Y.J.; Pulendran, B.; Palucka, K. Immunobiology of dendritic cells. Annu. Rev. Immunol. 2000, 18, 767–811. [Google Scholar] [CrossRef] [PubMed]
- Sprooten, J.; Ceusters, J.; Coosemans, A.; Agostinis, P.; De Vleeschouwer, S.; Zitvogel, L.; Kroemer, G.; Galluzzi, L.; Garg, A.D. Trial watch: dendritic cell vaccination for cancer immunotherapy. Oncoimmunology 2019, 8, e1638212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Vries, I.J.M.; Lesterhuis, W.J.; Scharenborg, N.M.; Engelen, L.P.H.; Ruiter, D.J.; Gerritsen, M.J.P.; Croockewit, S.; Britten, C.M.; Torensma, R.; Adema, G.J.; et al. Maturation of Dendritic Cells Is a Prerequisite for Inducing Immune Responses in Advanced Melanoma Patients. Clin. Cancer Res. 2003, 9, 5091–5100. [Google Scholar] [PubMed]
- McIlroy, D.; Gregoire, M. Optimizing dendritic cell-based anticancer immunotherapy: Maturation state does have clinical impact. Cancer Immunol. Immunother. 2003, 52, 583–591. [Google Scholar] [CrossRef]
- De Vries, I.J.M.; Krooshoop, D.J.E.B.; Scharenborg, N.M.; Lesterhuis, W.J.; Diepstra, J.H.S.; Van Muijen, G.N.P.; Strijk, S.P.; Ruers, T.J.; Boerman, O.C.; Oyen, W.J.G.; et al. Effective migration of antigen-pulsed dendritic cells to lymph nodes in melanoma patients is determined by their maturation state. Cancer Res. 2003, 63, 12–17. [Google Scholar]
- Saxena, M.; Balan, S.; Roudko, V.; Bhardwaj, N. Towards superior dendritic-cell vaccines for cancer therapy. Nat. Biomed. Eng. 2018, 2, 341–344. [Google Scholar] [CrossRef]
- Garg, A.D.; Coulie, P.G.; Van den Eynde, B.J.; Agostinis, P. Integrating Next-Generation Dendritic Cell Vaccines into the Current Cancer Immunotherapy Landscape. Trends Immunol. 2017, 38, 577–593. [Google Scholar] [CrossRef]
- Klebanoff, S.J.; Kettle, A.J.; Rosen, H.; Winterbourn, C.C.; Nauseef, W.M. Myeloperoxidase: a front-line defender against phagocytosed microorganisms. J. Leukoc. Biol. 2013, 93, 185–198. [Google Scholar] [CrossRef] [Green Version]
- Carrasco-Marín, E.; Paz-Miguel, J.E.; López-Mato, P.; Alvarez-Domínguez, C.; Leyva-Cobián, F. Oxidation of defined antigens allows protein unfolding and increases both proteolytic processing and exposes peptide epitopes which are recognized by specific T cells. Immunology 1998, 95, 314–321. [Google Scholar] [CrossRef]
- Alderman, C.J.J.; Shah, S.; Foreman, J.C.; Chain, B.M.; Katz, D.R. The role of advanced oxidation protein products in regulation of dendritic cell function. Free Radic. Biol. Med. 2002, 32, 377–385. [Google Scholar] [CrossRef]
- Marcinkiewicz, J.; Chain, B.M.; Olszowska, E.; Olszowski, S.; Zgliczyński, J.M. Enhancement of immunogenic properties of ovalbumin as a result of its chlorination. Int. J. Biochem. 1991, 23, 1393–1395. [Google Scholar] [CrossRef]
- Chiang, C.L.-L.; Ledermann, J.A.; Rad, A.N.; Katz, D.R.; Chain, B.M. Hypochlorous acid enhances immunogenicity and uptake of allogeneic ovarian tumor cells by dendritic cells to cross-prime tumor-specific T cells. Cancer Immunol. Immunother. 2006, 55, 1384–1395. [Google Scholar] [CrossRef] [PubMed]
- Prokopowicz, Z.M.; Arce, F.; Biedroń, R.; Chiang, C.L.-L.; Ciszek, M.; Katz, D.R.; Nowakowska, M.; Zapotoczny, S.; Marcinkiewicz, J.; Chain, B.M. Hypochlorous acid: a natural adjuvant that facilitates antigen processing, cross-priming, and the induction of adaptive immunity. J. Immunol. 2010, 184, 824–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcinkiewicz, J.; Olszowska, E.; Olszowski, S.; Zgliczynski, J.M. Enhancement of trinitrophenyl-specific humoral response to TNP proteins as the result of carrier chlorination. Immunology 1992, 76, 385–388. [Google Scholar]
- Chiang, C.L.-L.; Ledermann, J.A.; Aitkens, E.; Benjamin, E.; Katz, D.R.; Chain, B.M. Oxidation of ovarian epithelial cancer cells by hypochlorous acid enhances immunogenicity and stimulates T cells that recognize autologous primary tumor. Clin. Cancer Res. 2008, 14, 4898–4907. [Google Scholar] [CrossRef] [Green Version]
- Chiang, C.L.L.; Kandalaft, L.E.; Tanyi, J.; Hagemann, A.R.; Motz, G.T.; Svoronos, N.; Montone, K.; Mantia-Smaldone, G.M.; Smith, L.; Nisenbaum, H.L.; et al. A dendritic cell vaccine pulsed with autologous hypochlorous acid-oxidized ovarian cancer lysate primes effective broad antitumor immunity: From bench to bedside. Clin. Cancer Res. 2013, 19, 4801–4815. [Google Scholar] [CrossRef] [Green Version]
- Tanyi, J.L.; Bobisse, S.; Ophir, E.; Tuyaerts, S.; Roberti, A.; Genolet, R.; Baumgartner, P.; Stevenson, B.J.; Iseli, C.; Dangaj, D.; et al. Personalized cancer vaccine effectively mobilizes antitumor T cell immunity in ovarian cancer. Sci. Transl. Med. 2018, 10, eaao5931. [Google Scholar] [CrossRef] [Green Version]
- Biedroń, R.; Konopiński, M.K.; Marcinkiewicz, J.; Józefowski, S. Oxidation by neutrophils-derived HOCl increases immunogenicity of proteins by converting them into ligands of several endocytic receptors involved in antigen uptake by dendritic cells and macrophages. PLoS One 2015, 10, e0123293. [Google Scholar] [CrossRef] [Green Version]
- Chiang, C.L.L.; Hagemann, A.R.; Leskowitz, R.; Mick, R.; Garrabrant, T.; Czerniecki, B.J.; Kandalaft, L.E.; Powell, D.J.; Coukos, G. Day-4 myeloid dendritic cells pulsed with whole tumor lysate are highly immunogenic and elicit potent anti-tumor responses. PLoS One 2011, 6. [Google Scholar] [CrossRef]
- Chiang, C.L.-L.; Maier, D.A.; Kandalaft, L.E.; Brennan, A.L.; Lanitis, E.; Ye, Q.; Levine, B.L.; Czerniecki, B.J.; Powell Jr, D.J.; Coukos, G. Optimizing parameters for clinical-scale production of high IL-12 secreting dendritic cells pulsed with oxidized whole tumor cell lysate. J. Transl. Med. 2011, 9, 198. [Google Scholar] [CrossRef] [Green Version]
- Chong, C.; Marino, F.; Pak, H.; Racle, J.; Daniel, R.T.; Mü Ller, M.; Gfeller, D.; Coukos, G.; Bassani-Sternberg, M. High-throughput and sensitive immunopeptidomics platform reveals profound interferon γ-mediated remodeling of the human leukocyte antigen (HLA) ligandome. Mol. Cell. Proteomics 2018, 17, 533–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marino, F.; Chong, C.; Michaux, J.; Bassani-Sternberg, M. High-throughput, fast, and sensitive immunopeptidomics sample processing for mass spectrometry. Methods Mol. Biol. 2019, 1913, 67–79. [Google Scholar] [PubMed]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 2014, 13, 2513–2526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez, M.A.S.; Bassani-Sternberg, M.; Coukos, G.; Gfeller, D.; Zoete, V. Analysis of Secondary Structure Biases in Naturally Presented HLA-I Ligands. Front. Immunol. 2019, 10, 2731. [Google Scholar] [CrossRef]
- Goddard, T.D.; Huang, C.C.; Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018, 27, 14–25. [Google Scholar] [CrossRef]
- Sanner, M.F.; Olson, A.J.; Spehner, J.C. Reduced surface: An efficient way to compute molecular surfaces. Biopolymers 1996, 38, 305–320. [Google Scholar] [CrossRef]
- Bendell, C.J.; Liu, S.; Aumentado-Armstrong, T.; Istrate, B.; Cernek, P.T.; Khan, S.; Picioreanu, S.; Zhao, M.; Murgita, R.A. Transient protein-protein interface prediction: Datasets, features, algorithms, and the RAD-T predictor. BMC Bioinformatics 2014, 15, 82. [Google Scholar] [CrossRef] [Green Version]
- Kandalaft, L.E.; Chiang, C.L.; Tanyi, J.; Motz, G.; Balint, K.; Mick, R.; Coukos, G. A Phase I vaccine trial using dendritic cells pulsed with autologous oxidized lysate for recurrent ovarian cancer. J. Transl. Med. 2013, 11, 149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mookerjee Ananda; Graciotti Michele; Kandalaft Lana A cancer vaccine with dendritic cells differentiated with GM-CSF and IFNa and pulsed with a squaric acid treated cell lysate improves T cell priming and tumor growth control in a mouse model. BioImpacts 2018, 8, 211–221. [CrossRef] [PubMed]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef] [PubMed]
- Pattison, D.I.; Davies, M.J. Absolute rate constants for the reaction of hypochlorous acid with protein side chains and peptide bonds. Chem. Res. Toxicol. 2001, 14, 1453–1464. [Google Scholar] [CrossRef]
- Hawkins, C.L.; Pattison, D.I.; Davies, M.J. Hypochlorite-induced oxidation of amino acids, peptides and proteins. Amino Acids 2003, 25, 259–274. [Google Scholar] [CrossRef]
- Ehrenshaft, M.; Deterding, L.J.; Mason, R.P. Tripping up Trp: Modification of protein tryptophan residues by reactive oxygen species, modes of detection, and biological consequences. Free Radic. Biol. Med. 2015, 89, 220–228. [Google Scholar] [CrossRef] [Green Version]
- Luo, S.; Levine, R.L. Methionine in proteins defends against oxidative stress. FASEB J. 2009, 23, 464–472. [Google Scholar] [CrossRef] [Green Version]
- Kowaltowski, A.J.; Vercesi, A.E. Mitochondrial damage induced by conditions of oxidative stress. Free Radic. Biol. Med. 1999, 26, 463–471. [Google Scholar] [CrossRef]
- Guo, C.Y.; Sun, L.; Chen, X.P.; Zhang, D.S. Oxidative stress, mitochondrial damage and neurodegenerative diseases. Neural Regen. Res. 2013, 8, 2003–2014. [Google Scholar] [CrossRef]
- Kalmar, B.; Greensmith, L. Induction of heat shock proteins for protection against oxidative stress. Adv. Drug Deliv. Rev. 2009, 61, 310–318. [Google Scholar] [CrossRef]
- Luo, M.; Joiner, M.L.A. Stress response signaling pathways may lead to mitochondrial biogenesis. Diabetes 2014, 63, 1831–1832. [Google Scholar] [CrossRef] [Green Version]
- Nishinaka, Y.; Masutani, H.; Nakamura, H.; Yodoi, J. Regulatory roles of thioredoxin in oxidative stress-induced cellular responses. Redox Rep. 2004, 6, 289–295. [Google Scholar] [CrossRef]
- Niforou, K.; Cheimonidou, C.; Trougakos, I.P. Molecular chaperones and proteostasis regulation during redox imbalance. Redox Biol. 2014, 2, 323–332. [Google Scholar] [CrossRef] [Green Version]
- Rogers, K.R.; Morris, C.J.; Blake, D.R. Oxidation of thiol in the vimentin cytoskeleton. Biochem. J. 1991, 275, 789–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landino, L.M.; Hagedorn, T.D.; Kim, S.B.; Hogan, K.M. Inhibition of tubulin polymerization by hypochlorous acid and chloramines. Free Radic. Biol. Med. 2011, 50, 1000–1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Liu, L.; Jin, L.; Yi, X.; Dang, E.; Yang, Y.; Li, C.; Gao, T. Oxidative stress-induced calreticulin expression and translocation: New insights into the destruction of melanocytes. J. Invest. Dermatol. 2014, 134, 183–191. [Google Scholar] [CrossRef] [Green Version]
- Shimi, T.; Goldman, R.D. Nuclear lamins and oxidative stress in cell proliferation and longevity. Adv. Exp. Med. Biol. 2014, 773, 415–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakajima, H.; Itakura, M.; Kubo, T.; Kaneshige, A.; Harada, N.; Izawa, T.; Azuma, Y.T.; Kuwamura, M.; Yamaji, R.; Takeuchi, T. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) aggregation causes mitochondrial dysfunction during oxidative stress-induced cell death. J. Biol. Chem. 2017, 292, 4727–4742. [Google Scholar] [CrossRef] [Green Version]
- Farah, M.E.; Sirotkin, V.; Haarer, B.; Kakhniashvili, D.; Amberg, D.C. Diverse protective roles of the actin cytoskeleton during oxidative stress. Cytoskeleton 2011, 68, 340–354. [Google Scholar] [CrossRef] [Green Version]
- Ferrington, D.A.; Gregerson, D.S. Immunoproteasomes: Structure, function, and antigen presentation. Prog. Mol. Biol. Trans. Sci. 2012, 109, 75–112. [Google Scholar]
- Blum, J.S.; Wearsch, P.A.; Cresswell, P. Pathways of Antigen Processing. Annu. Rev. Immunol. 2013, 31, 443–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bassani-Sternberg, M.; Pletscher-Frankild, S.; Jensen, L.J.; Mann, M. Mass Spectrometry of Human Leukocyte Antigen Class I Peptidomes Reveals Strong Effects of Protein Abundance and Turnover on Antigen Presentation. Mol. Cell. Proteomics 2015, 14, 658–673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreatta, M.; Lund, O.; Nielsen, M. Simultaneous alignment and clustering of peptide data using a Gibbs sampling approach. Bioinformatics 2013, 29, 8–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rapin, N.; Hoof, I.; Lund, O.; Nielsen, M. MHC motif viewer. Immunogenetics 2008, 60, 759–765. [Google Scholar] [CrossRef]
- Andersen, M.H.; Svane, I.; Becker, J.C.; Straten, P.T. The universal character of the tumor-associated antigen survivin. Clin. Cancer Res. 2007, 13, 5991–5994. [Google Scholar] [CrossRef] [Green Version]
- Wargo, J.A.; Robbins, P.F.; Li, Y.; Zhao, Y.; El-Gamil, M.; Caragacianu, D.; Zheng, Z.; Hong, J.A.; Downey, S.; Schrump, D.S.; et al. Recognition of NY-ESO-1+ tumor cells by engineered lymphocytes is enhanced by improved vector design and epigenetic modulation of tumor antigen expression. Cancer Immunol. Immunother. 2009, 58, 383–394. [Google Scholar] [CrossRef] [Green Version]
- Weon, J.L.; Potts, P.R. The MAGE protein family and cancer. Curr. Opin. Cell Biol. 2015, 37, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Sabado, R.L.; Balan, S.; Bhardwaj, N. Dendritic cell-based immunotherapy. Cell Res. 2017, 27, 74–95. [Google Scholar] [CrossRef] [Green Version]
- Pullar, J.M.; Vissers, M.C.; Winterbourn, C.C. Living with a killer: the effects of hypochlorous acid on mammalian cells. IUBMB Life 2000, 50, 259–266. [Google Scholar] [CrossRef]
- Levine, R.L.; Mosoni, L.; Berlett, B.S.; Stadtman, E.R. Methionine residues as endogenous antioxidants in proteins. Proc. Natl. Acad. Sci. USA 1996, 93, 15036–15040. [Google Scholar] [CrossRef] [Green Version]
- Moskovitz, J. Methionine sulfoxide reductases: Ubiquitous enzymes involved in antioxidant defense, protein regulation, and prevention of aging-associated diseases. Biochim. Biophys. Acta Proteins Proteom. 2005, 1703, 213–219. [Google Scholar] [CrossRef]
- Zhang, X.H.; Weissbach, H. Origin and evolution of the protein-repairing enzymes methionine sulphoxide reductases. Biol. Rev. 2008, 83, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Binder, R.J.; Suto, R.; Anderson, K.M.; Srivastava, P.K. Necrotic but not apoptotic cell death releases heat shock proteins, which deliver a partial maturation signal to dendritic cells and activate the NF-κB pathway. Int. Immunol. 2000, 12, 1539–1546. [Google Scholar] [CrossRef] [PubMed]
- Somersan, S.; Larsson, M.; Fonteneau, J.F.; Basu, S.; Srivastava, P.; Bhardwaj, N. Primary Tumor Tissue Lysates Are Enriched in Heat Shock Proteins and Induce the Maturation of Human Dendritic Cells. J. Immunol. 2001, 167, 4844–4852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coelho, V.; Faria, A.M.C. HSP60: Issues and insights on its therapeutic use as an immunoregulatory agent. Front. Immunol. 2012, 2, 97. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zeng, X.; He, L.; Yuan, H. Dendritic cell activation and maturation induced by recombinant calreticulin fragment 39-272. Int. J. Clin. Exp. Med. 2015, 8, 7288–7296. [Google Scholar]
- Liu, X.; Li, J.; Liu, Y.; Ding, J.; Tong, Z.; Liu, Y.; Zhou, Y.; Liu, Y. Calreticulin acts as an adjuvant to promote dendritic cell maturation and enhances antigen-specific cytotoxic T lymphocyte responses against non-small cell lung cancer cells. Cell. Immunol. 2016, 300, 46–53. [Google Scholar] [CrossRef]
- Srivastava, P. Roles of heat-shock proteins in innate and adaptive immunity. Nat. Rev. Immunol. 2002, 2, 185–194. [Google Scholar] [CrossRef]
- Shevtsov, M.; Multhoff, G. Heat shock protein-Peptide and HSP-based immunotherapies for the treatment of cancer. Front. Immunol. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Baldin, A.V.; Zamyatnin, A.A., Jr.; Bazhin, A.V.; Xu, W.-H.; Savvateeva, L.V. Advances in the Development of Anticancer HSP-based Vaccines. Curr. Med. Chem. 2018, 26, 427–445. [Google Scholar] [CrossRef]
- Kreiter, S.; Vormehr, M.; Van De Roemer, N.; Diken, M.; Löwer, M.; Diekmann, J.; Boegel, S.; Schrörs, B.; Vascotto, F.; Castle, J.C.; et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature 2015, 520, 692–696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melief, C.J.; van der Burg, S.H. Immunotherapy of established (pre)malignant disease by synthetic long peptide vaccines. Nat. Rev. Cancer 2008, 8, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Alspach, E.; Lussier, D.M.; Miceli, A.P.; Kizhvatov, I.; DuPage, M.; Luoma, A.M.; Meng, W.; Lichti, C.F.; Esaulova, E.; Vomund, A.N.; et al. MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature 2019, 574, 696–701. [Google Scholar] [CrossRef] [PubMed]
- Boegel, S.; Löwer, M.; Bukur, T.; Sahin, U.; Castle, J.C. A catalog of HLA type, HLA expression, and neoepitope candidates in human cancer cell lines. Oncoimmunology 2014, 3, e954893. [Google Scholar] [CrossRef] [PubMed]








© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Graciotti, M.; Marino, F.; Pak, H.; Baumgaertner, P.; Thierry, A.-C.; Chiffelle, J.; Perez, M.A.S.; Zoete, V.; Harari, A.; Bassani-Sternberg, M.; et al. Deciphering the Mechanisms of Improved Immunogenicity of Hypochlorous Acid-Treated Antigens in Anti-Cancer Dendritic Cell-Based Vaccines. Vaccines 2020, 8, 271. https://doi.org/10.3390/vaccines8020271
Graciotti M, Marino F, Pak H, Baumgaertner P, Thierry A-C, Chiffelle J, Perez MAS, Zoete V, Harari A, Bassani-Sternberg M, et al. Deciphering the Mechanisms of Improved Immunogenicity of Hypochlorous Acid-Treated Antigens in Anti-Cancer Dendritic Cell-Based Vaccines. Vaccines. 2020; 8(2):271. https://doi.org/10.3390/vaccines8020271
Chicago/Turabian StyleGraciotti, Michele, Fabio Marino, HuiSong Pak, Petra Baumgaertner, Anne-Christine Thierry, Johanna Chiffelle, Marta A. S. Perez, Vincent Zoete, Alexandre Harari, Michal Bassani-Sternberg, and et al. 2020. "Deciphering the Mechanisms of Improved Immunogenicity of Hypochlorous Acid-Treated Antigens in Anti-Cancer Dendritic Cell-Based Vaccines" Vaccines 8, no. 2: 271. https://doi.org/10.3390/vaccines8020271
APA StyleGraciotti, M., Marino, F., Pak, H., Baumgaertner, P., Thierry, A. -C., Chiffelle, J., Perez, M. A. S., Zoete, V., Harari, A., Bassani-Sternberg, M., & Kandalaft, L. E. (2020). Deciphering the Mechanisms of Improved Immunogenicity of Hypochlorous Acid-Treated Antigens in Anti-Cancer Dendritic Cell-Based Vaccines. Vaccines, 8(2), 271. https://doi.org/10.3390/vaccines8020271