Coronavirus Disease (COVID-19) Control between Drug Repurposing and Vaccination: A Comprehensive Overview
Abstract
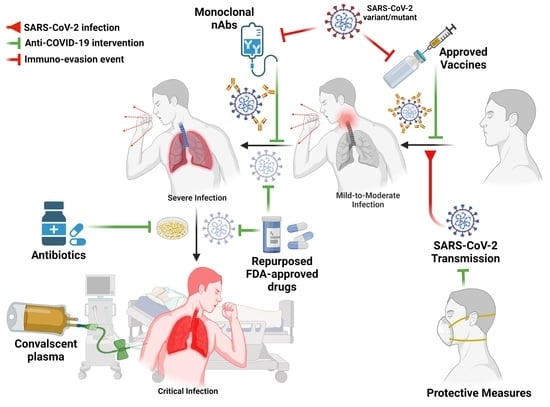
:1. Introduction
2. SARS-CoV-2 Life Cycle and Its Potential Targets
3. Clinical Management of COVID-19 Patients
3.1. Mild “Symptomatic Treatment”
3.2. Moderate “Pneumonia Treatment”
3.3. Severe and Critical Symptoms
4. Prophylactic and Therapeutic Interventions
4.1. SARS-CoV-2 Vaccine Candidates
| Approach | Vaccine Name | Vaccine Class | Manufacturer | Efficacy | Dosing and Storage |
|---|---|---|---|---|---|
| mRNA vaccine | mRNA- 1273 | Encapsulated mRNA | Moderna/NIAID | 94.1% against original strain | 2 doses- 4 weeks apart Stored at −20 °C |
| BNT162b2 | Encapsulated mRNA | BioNTech/Pfizer/Fosun Pharma | 95% against original strain | 2 doses- 3 weeks apart Stored at −70 °C | |
| Replication of defective viral-vector vaccine | Ad5-nCoV | Viral vector | CanSino Biological in collaboration with Beijing Institute of Biotechnology and Academy of Military Medical Sciences | Phase III (ongoing) | 1 dose- Stored at 2–8 °C |
| ChAdOx1 /AZD1222 | Viral vector | Oxford University/AstraZeneca | 70.4% against original strain | 2 doses- 4 weeks apart Stored at 2–8 °C | |
| Sputnik-V /Gam-COVID-Vac | Viral vector | Acellena Contract Drug Research and Development in collaboration with Gamaleya Research Institute and Health Ministry of the Russian Federation | 91.4% against original strain | 2 doses- 3 weeks apart Stored at 2–8 °C | |
| JNJ-78436735/ Ad26.COV2.S | Viral vector | Johnson & Johnson | 72% against original strain | 1 dose- Stored at 2–8 °C | |
| Inactivated vaccine | CoronaVac “ Sinovac” | Inactivated virus | Sinovac Research and Development Co. | 50% against original strain | 2 doses- 2 weeks apart Stored at 2–8 °C |
| BBIBP-CorV | Inactivated virus | Beijing Institute of Biotechnology | 79.34% against original strain | 2 doses- 3 weeks apart Stored at 2–8 °C | |
| Sinopharm (Wuhan) | Inactivated virus | China National Pharmaceutical Group (Sinopharm) in collaboration with Wuhan Institute of Biological Products | undisclosed | 2 doses- 3 weeks apart Stored at 2–8 °C | |
| BBV152/ Covaxin | Inactivated virus | Bharat Biotech | 81% against original strain | 2 doses- 4 weeks apart Stored at 2–8 °C | |
| Subunit vaccine | NVX- CoV2373 | Recombinant spike (rS) and Matrix-M1 proteins | Novavax | 96% against original strain | 2 doses- 3 weeks apart Stored at 2–8 °C |
| ZF2001 | The repeated dimeric form of RBD of the SARS-CoV-2 S protein | Anhui Zhifei Longcom/ Chinese Academy of Medicine | 92–97% | 3 doses- 4 weeks apart Stored at 2–8 °C | |
| Virus-like particle (VLP) | CoVLP (NCT04450004) | plant-produced VLP vaccine candidate expressing SARS-CoV-2 spike protein | Medicago/GlaxoSmithKline | Phase II–III clinical trial (ongoing) | 2 doses- 3 weeks apart Stored at 2–8 °C |
4.2. Immune Response to Coronavirus Infection
4.3. Immune-Evasion of SARS-CoV-2 and Its Emerging Variants
5. Available Therapeutic Interventions against SARS-CoV-2
5.1. Chloroquine and Hydroxychloroquine
Clinical Trials
- (a)
- In a randomized, placebo-controlled, and double-blind clinical trial including 821 adult participants from U.S.A. and Canada, the volunteers were in direct exposure to COVID-19 patients without any safety measures (neither face masks nor shields). On the fourth day after exposure, the participants were randomized to a placebo control group or hydroxychloroquine group. Laboratory-confirmed COVID-19 infection within 14 days was the primary outcome of the trial. Unfortunately, this clinical trial advised against the prescription of hydroxychloroquine as a postexposure prophylactic intervention for COVID-19 [150].
- (b)
- A meta-analysis of ongoing, completed, or discontinued randomized clinical trials on the use of hydroxychloroquine or chloroquine to treat COVID-19 patients was conducted. The study ended up with a conclusion that the treatment with hydroxychloroquine is associated with increased mortality among COVID-19 patients [151].
- (c)
- In a placebo-controlled, randomized, blind clinical trial at 34 hospitals in the U.S.A., 479 COVID-19 patients were randomized and treated either with placebo or hydroxychloroquine for 14 days. The outcomes of this randomized trial advised against the use of hydroxychloroquine in the treatment protocol of hospitalized COVID-19 adults [152].
5.2. Antiparasitic Drug
5.2.1. Ivermectin
Clinical Trials
- (a)
- Potential study participants were chosen at random from the state’s electronic database of patients with symptomatic, laboratory-confirmed COVID-19 during the study period. The investigation was completed by 398 (99.5%) of 400 patients including 231 women (58%) (median age 37 years) in the primary analysis population. The median time to symptoms clearance in the ivermectin group was 10 days, while the placebo group took 12 days [156]. Three weeks posttreatment, 82% of individuals using ivermectin had gotten rid of their symptoms, while 79% of those taking placebo had. The most common side effect was a headache, which was observed by 104 patients (52%) who received ivermectin and 111 patients (56%) who received a placebo [156].
- (b)
- In another clinical trial, a total of 32 COVID-19 patients were randomized to receive the standard of care (SOC) treatment with variable doses of ivermectin (100–400 mcg/kg). Interestingly, SOC treatment plus ivermectin showed high safety and could significantly reduce symptoms and viral loads in all patients within 7 days [157].
5.2.2. Nitazoxanide (NTZ)
Clinical Trials
- (a)
- Early-stage 392 symptomatic COVID-19 patients were randomized to receive NTZ (196 cases, dose: 500 mg/3 times daily/5 days) against the SOC treatment (196 cases, placebo) [161]. Interestingly, 11% improvement in symptom-free days in NTZ-treated patients was documented compared to the placebo group [161].
- (b)
- From 20 May to 21 September 2020, a randomized clinical trial comparing NTZ (600 mg, twice) against placebo for seven days in 50 COVID-19 patients with mild respiratory insufficiency. Interestingly, a decrease in the time for hospital discharge, faster clinical swab negativity, and a significant reduction in the levels of inflammatory and lymphocyte T cells activation markers were documented among NTZ-treated patients compared to placebo [162].
5.2.3. Niclosamide
Clinical Trials
- (a)
- From 29 June to 8 August 2020, a total of 34 healthy individuals in Denmark were given a niclosamide formulation “UNI91104”, and ten were given a placebo in a placebo-controlled clinical trial. The results of this study showed that the UNI91104 is a promising and safe anti-COVID-19 drug candidate following intranasal administration [168].
- (b)
- Another study comprised 75 COVID-19 patients who received SOC plus niclosamide in the experimental group and 75 COVID-19 patients who received only SOC therapy as a control group [169]. Within 30 days of follow-up, there was no significant difference in the incidence of death versus recovery between the two research groups. The niclosamide supplement group’s median survival time to cure was considerably shorter than the control group. After adjusting for comorbidity count, niclosamide add-on treatment increased the chance of cure by 60% each day compared to the control group [169].
5.3. Antibiotics
5.3.1. Azithromycin
Clinical Trials
- (a)
- In a clinical trial, 298 enrolled COVID-19-positive individuals between 3 June 2020 to 29 January 2021 were split into two groups: 145 were given azithromycin together with the SOC treatment, and 147 were given the SOC treatment alone [172]. This study ended up with the conclusion that, in mild-to-moderate COVID-19 patients who were managed without hospitalization, adding azithromycin to the SOC treatment did not diminish the risk of subsequent hospital admission or death.
- (b)
- From 22 May and 30 November 2020, 2265 COVID-19 positive participants were divided into three groups: azithromycin plus SOC treatment (540 patients), SOC treatment (875 patients), and a third group that received other interventions (850 patients). The outcome of this clinical trial advised against the routine prescription of azithromycin for shortening the recovery time or reducing the risk of hospitalization for suspected COVID-19 cases [173]
- (c)
- Through May 2020 to March 2021, 263 outpatients with SARS-CoV-2 infection were randomized into azithromycin group (n = 171) or placebo group (n = 92). Compared to the placebo group, treatment with a single dose of azithromycin (500 mg once daily for 14 days) did not lead to faster relief of the symptoms. To this point, the study did not recommend the routine use of azithromycin for COVID-19 outpatients [174].
5.3.2. Fluoroquinolones
Clinical Trials
- (a)
- From 15 February to 15 March 2020, 94 patients with COVID-19 including 27 severe patients at the Intensive Care Unit (ICU) and 74 ordinary patients at the general isolation ward in Wuhan Xiehe Hospital were treated with the anti-influenza drug arbidol (100 mg, three times daily for 14 days) and moxifloxacin (400 mg daily for 7–14 days) [177]. The study ended up with a conclusion that arbidol and moxifloxacin could reduce viral load and inflammation in COVID-19 patients
- (b)
- Between 20 January and 15 March 2020, a number of 55 COVID-19 patients with mild-to-severe symptoms were hospitalized at Shenyang Sixth People’s Hospital. The treatment protocol in 53 patients included antiviral umifenovir and lopinavir/ritonavir therapies. A total of 29 patients were administered antibiotics, including moxifloxacin or linezolid. Moreover, 7 patients were treated with glucocorticoids and 9 with immunomodulators. All patients recovered, and this can partially emphasize the prophylactic administration of common antibiotics to reduce the risk of the fatal secondary bacterial co-infection [178] and their potential anti-SARS-CoV-2 activity.
5.4. Broad Spectrum Antivirals
5.4.1. Triazavirin (TZV)
Clinical Trial
5.4.2. Umifenovir
Clinical Trial
5.5. RNA-Dependent RNA Polymerase (RdRp) Inhibitors
5.5.1. Favipiravir (FPV)
Clinical Trial
5.5.2. Remdesivir
Clinical Trial
5.5.3. Molnupiravir
Clinical Trial
5.6. Protease Inhibitors
5.6.1. Danoprevir
Clinical Trial
5.6.2. Darunavir
Clinical Trial
5.7. Nucleoside Inhibitors
5.7.1. Azvudine
Clinical Trial
5.7.2. Tenofovir Disoproxil Fumarate
Clinical Trial
5.7.3. Ribavirin
Clinical Trial
5.8. Antihypertensive Drugs
Clinical Trials
- (a)
- A total of 50,615 patients were included in 40 studies (21 cross-sectional, 2 case-control, and 17 cohorts). In subgroups by study design and considering adjusted effects, the use of ACEIs or ARBs was not linked with all-cause mortality. Disease severity was linked to the use of an ACEI or an ARB. There were no significant links discovered between the use of ACEIs or ARBs and hospital discharge, hospitalization, mechanical ventilation, length of stay, or biomarkers [204].
- (b)
- Between 28 February and 18 August 2020, data from 228,722 veterans with a history of hypertension who had COVID-19 testing were reviewed to see if they were taking ACEIs or ARBs and the possible impact on (1) a positive COVID-19 test and (2) a severe outcome (hospitalization, mortality, and use of the intensive care unit (ICU) and/or mechanical ventilation). Interestingly, data analysis showed that ACEI use reduced the likelihood of a positive COVID-19 test in veterans with hypertension. When comparing COVID-19 inpatients to outpatients, the use of ACEI, but not ARB, was linked with a significantly higher likelihood of using mechanical ventilators [205].
- (c)
- In this retrospective observational study, 681 COVID-19 patients who were admitted to Sina Hospital in Tehran, Iran, from 20 February to 29 May 2020, were investigated. A total of 37 were eliminated due to inadequate medical records, and 8 utilized ACEIs, leaving 636 patients in the study. In this group, 108 (17.0%) patients died, and 407 (64.0%) patients developed severe COVID-19. ARBs were given to 122 (48.0%) of the 254 individuals with hypertension (39.9%). The study found no independent connection between taking ARBs and in-hospital outcomes, except for acute kidney damage (AKI), after adjusting for known confounders, in patients with confirmed or clinically suspected COVID-19, hypertension, or nonhypertensive. The study discovered that stopping ARBs while in the hospital was linked to a higher risk of death [206].
- (d)
- The trial enrolled 659 patients from 29 different locations around Brazil. All of the subjects were taking an ACE inhibitor or an ARB for a long time and were hospitalized with COVID-19. Patients were randomly assigned to either discontinue taking the ACEI/ARB for 30 days or continue taking it. The average number of days alive and out of the hospital for patients who stopped taking ACEI/ARBs was 21.9 days, while the average number of days alive and out of hospital for patients who remained taking these medications was 22.9 days. Between the suspending and continuing groups, the average ratio of days alive and out of the hospital was 0.95 [207]. By the end of 30 days, 91.8% of patients in the suspending ACEI/ARB group were alive and out of the hospital, compared to 95% in the continuing group [207].
- (e)
- Another retrospective cohort study of veterans comparing the use of ARB/ACEI versus non-ARB/ACEI and ARB versus ACEI among COVID-19 outpatients and hospitalized inpatients. This observational study recommended the continuous usage of ARB or ACEI during treatment regimen for those hypertensive patients who already used them before COVID-19 infection [208].
- (f)
- A large cohort study (8.3 million people) comprised 19486 COVID-19 patients (20–99 years old) and 1286 ICU patients. The study data showed that the ACEI and ARBs treatment was associated with reduced risks of COVID-19 disease in studied populations after adjustment of a wide range of variables. Nevertheless, the ACEI and ARBs did not reduce the risks of receiving ICU care. Interestingly, ethnic-specific effects of ACEI/ARBs on COVID-19 disease susceptibility and severity were observed [209].
5.9. Antiviral against Influenza Viruses
5.9.1. Oseltamivir
Clinical Trial
- (a)
- Patients who presented with influenza-like illness (ILI) and suspected/confirmed positive for coronavirus were randomized to receive either standard therapy or standard care plus oseltamivir [210]. The most important outcome was time to recovery, which was defined as being able to resume normal activities with moderate to no fever, headache, or muscle soreness.
- (b)
- Coronaviruses were discovered in 308 (9%) of the 3266 participants in the randomized experiment; 153 were given usual care, and 155 were given usual care plus oseltamivir. The primary result was determined in 136 and 147 people, respectively. Patients given oseltamivir had a faster recovery time: 4 days (interquartile range (IQR) 3–6) versus 5 days (IQR 3–8; hazard ratio 1.31; 95 percent confidence interval = 1.03 to1.66; p = 0.026) [210].
5.9.2. Baloxavir Marboxil
Clinical Trials
5.10. Immunomodulators and Neutralizing Antibodies
5.10.1. Tocilizumab
Clinical Trials
- (a)
- (b)
- The first patient with multiple myeloma got a single dose of intravenous tocilizumab (8 mg/kg) on day 9 of hospitalization. He was given 40 mg methylprednisolone for four days before receiving tocilizumab. Despite improvements in breathing, chest tightness and CT imaging did not improve. After treatment with tocilizumab, the IL-6 blood level dropped from 122 to 21 pg/mL on day 18, and clinical symptoms and chest CT imaging both improved [216].
- (c)
- The researchers conducted a retrospective single-center case study of 21 Chinese patients with critical (19%) and severe (81%) COVID19 infection. COVID-19 was defined as a condition requiring mechanical ventilation or organ support in the intensive care unit [217]. Tachypnea and/or respiratory failure were common in patients with severe COVID-19. The average age of the patients was 56.8 16.5 years, with 85.7% of them being men. The concentration of IL-6 was 132.4 × 278.5 pg/mL on average. All of the patients were given lopinavir, methylprednisolone, a variety of symptom relievers, and oxygen therapy. In addition to regular therapy, all patients got a single dose of intravenous tocilizumab 400 mg, and three patients received a second dose of tocilizumab 400 mg after a 12-hour break. It is worth noting that all of the patients had undergone normal treatment seven days before receiving tocilizumab; yet, there was no change in symptoms, hypoxemia, or CT imaging [217]. Tocilizumab treatment resulted in an immediate reduction in symptoms, CT opacity alterations, and hypoxemia. Within 24 h of receiving the medication, the patients’ fevers had entirely subsided. Radiological improvement in ground-glass opacities was seen in 91% of patients. The blood and oxygenation levels of 19 people were measured. The mean C-reactive protein (CRP) level reduced from 75.1 66.8 to 2.72 3.6 mg/mL on day 5 after medication [217]. Furthermore, within 5 days of treatment, patients’ oxygen saturations improved statistically considerably: 1 patient no longer required supplemental oxygen, 15 required less oxygen, 1 started the ventilator weaning process, and 2 were extubated. Finally, 19 people were discharged, with 2 remaining in the hospital in stable condition [217].
- (d)
- Another retrospective single-center case study was conducted in 15 Chinese patients with COVID-19 who were moderately unwell (13.3%), seriously ill (40%), and critically ill (46.7%). Patients were, on average, 73 years old, with a 75% male preponderance. Diabetes mellitus, hypertension, and a prior cerebrovascular accident were among the patients’ baseline comorbidities in 27%, 60%, and 20%, respectively. All patients received tocilizumab (80600 mg) at least once, either alone (47%) or in combination with methylprednisolone (53%) [218]. Tocilizumab was administered in three doses to 33 percent of the patients. By the seventh day after treatment, 67% of patients were clinically stable, 13% had deteriorated disease, and 20% had died [218].
- (e)
- In COVID-19, the studies also compared tocilizumab to standard care or alternative medicines that may have a favorable effect. Finally, main study endpoints such as death rate, remission of fever after 24 h, clinical improvement, biochemical response, oxygen saturation, need for mechanical ventilation, and change in Sequential Organ Failure Assessment (SOFA) score revealed significant variability [219].
- (f)
- According to China’s National Health Commission, tocilizumab should be investigated for the treatment of COVID-19 infected people who have high IL-6 levels and substantial lung damage [220]. Tocilizumab’s adverse effects include infection, increased serum cholesterol, ALT, AST, and injection site sensitivity. Understanding the safety of tocilizumab and the potential for harm in different disorders could help clinicians determine potential COVID-19 treatment exclusion criteria. Tocilizumab has several contraindications, including known hypersensitivity to the medicine and active infection (including localized infection). Herpes zoster reactivation has been observed [221,222,223].
5.10.2. Interferon β-1α
Clinical Trial
5.10.3. Baricitinib
Clinical Trials
5.10.4. Corticosteroid Therapy
5.10.5. Convalescent Plasma
Clinical Trial
5.10.6. Neutralizing Monoclonal Antibodies (mAbs) Cocktails
Clinical Trials
- (a)
- In a randomized phase 3 clinical trial, 1035 mild-to-moderate COVID-19 patients with a high risk of severe disease development were randomized to receive either a single intravenous infusion of placebo or bamlanivimab–etesevimab mAb combination. Interestingly, the mAb cocktail resulted in an accelerated decline of SARS-CoV-2 viral loads and reduced rates of hospitalization and death.
- (b)
- In a retrospective cohort study, 696 individuals with mild-to-moderate symptoms and treated with casirivimab–imdevimab mAb combination were compared to the untreated control group “696 mild-to-moderate COVID-19 patients” in different sites in the U.S.A. The study ended up with a conclusion that casirivimab–imdevimab mAb combination led to a significant reduction in the hospitalization rate of high-risk COVID-19 patients when compared to the control group [243].
- (c)
- In a randomized, multicenter, phase 3 clinical trial, 583 high-risk mild-to-moderate COVID-19 patients were randomized to have 292 individuals in the control group “placebo” and 291 in sotrovimab group. At 29 days postrandomization, the study was ended and resulted in the recommendation that the sotrovimab reduced the COVID-19 progression in studied high-risk patients [244].
5.11. Supplementary Treatments
5.11.1. Oxygen Therapy
5.11.2. Herbal Therapy
5.11.3. Zinc and Vitamin Supplements
6. Future Perspective and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Jong, J.C.; Claas, E.; Osterhaus, A.D.; Webster, R.G.; Lim, W. A pandemic warning? Nature 1997, 389, 554. [Google Scholar] [CrossRef]
- El Gizawy, H.A.; Boshra, S.A.; Mostafa, A.; Mahmoud, S.H.; Ismail, M.I.; Alsfouk, A.A.; Taher, A.T.; Al-Karmalawy, A.A. Pimenta dioica (L.) Merr. Bioactive Constituents Exert Anti-SARS-CoV-2 and Anti-Inflammatory Activities: Molecular Docking and Dynamics, In Vitro, and In Vivo Studies. Molecules 2021, 26, 5844. [Google Scholar] [CrossRef]
- Gonzalez, J.; Gomez-Puertas, P.; Cavanagh, D.; Gorbalenya, A.; Enjuanes, L. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch. Virol. 2003, 148, 2207–2235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, A.M.; Lefkowitz, E.; Adams, M.J.; Carstens, E.B. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses; Elsevier: Amsterdam, The Netherlands, 2011; Volume 9. [Google Scholar]
- Mostafa, A.; Kandeil, A.; Shehata, M.; El Shesheny, R.; Samy, A.M.; Kayali, G.; Ali, M.A. Middle east respiratory syndrome coronavirus (mers-cov): State of the science. Microorganisms 2020, 8, 991. [Google Scholar] [CrossRef] [PubMed]
- Zaki, A.M.; Van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.; Fouchier, R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef]
- Hui, D.S.; Zumla, A. Severe acute respiratory syndrome: Historical, epidemiologic, and clinical features. Infect. Dis. Clin. 2019, 33, 869–889. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Xu, Y.; Bao, L.; Zhang, L.; Yu, P.; Qu, Y.; Zhu, H.; Zhao, W.; Han, Y.; Qin, C. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses 2019, 11, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kandeil, A.; Mostafa, A.; El-Shesheny, R.; Shehata, M.; Roshdy, W.H.; Ahmed, S.S.; Gomaa, M.; Taweel, A.E.; Kayed, A.E.; Mahmoud, S.H. Coding-complete genome sequences of two sars-cov-2 isolates from egypt. Microbiol. Resour. Announc. 2020, 9, e00489-20. [Google Scholar] [CrossRef]
- Zhang, L.; Shen, F.M.; Chen, F.; Lin, Z. Origin and Evolution of the 2019 Novel Coronavirus. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2020, 71, 882–883. [Google Scholar] [CrossRef] [Green Version]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Wrobel, A.G.; Benton, D.J.; Xu, P.; Roustan, C.; Martin, S.R.; Rosenthal, P.B.; Skehel, J.J.; Gamblin, S.J. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat. Struct. Mol. Biol. 2020, 27, 763–767. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.-L.; Wang, Y.-M.; Wu, Z.-Q.; Xiang, Z.-C.; Guo, L.; Xu, T.; Jiang, Y.-Z.; Xiong, Y.; Li, Y.-J.; Li, X.-W.; et al. Identification of a novel coronavirus causing severe pneumonia in human: A descriptive study. Chin. Med. J. 2020, 133, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Latinne, A.; Hu, B.; Olival, K.J.; Zhu, G.; Zhang, L.; Li, H.; Chmura, A.A.; Field, H.E.; Zambrana-Torrelio, C.; Epstein, J.H.; et al. Origin and cross-species transmission of bat coronaviruses in China. Nat. Commun. 2020, 11, 4235. [Google Scholar] [CrossRef]
- Islam, M.S.; Sazzad, H.M.S.; Satter, S.M.; Sultana, S.; Hossain, M.J.; Hasan, M.; Rahman, M.; Campbell, S.; Cannon, D.L.; Ströher, U.; et al. Nipah Virus Transmission from Bats to Humans Associated with Drinking Traditional Liquor Made from Date Palm Sap, Bangladesh, 2011–2014. Emerg. Infect. Dis. 2016, 22, 664–670. [Google Scholar] [CrossRef] [Green Version]
- Xiao, X.; Newman, C.; Buesching, C.D.; Macdonald, D.W.; Zhou, Z.-M. Animal sales from Wuhan wet markets immediately prior to the COVID-19 pandemic. Sci. Rep. 2021, 11, 11898. [Google Scholar] [CrossRef]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Pei, S.; Chen, B.; Song, Y.; Zhang, T.; Yang, W.; Shaman, J. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2). Science 2020, 368, 489–493. [Google Scholar] [CrossRef] [Green Version]
- Oran, D.P.; Topol, E.J. Prevalence of Asymptomatic SARS-CoV-2 Infection: A Narrative Review. Ann. Intern. Med. 2020, 173, 362–367. [Google Scholar] [CrossRef]
- Zaki, N.; Mohamed, E.A. The estimations of the COVID-19 incubation period: A scoping reviews of the literature. J. Infect. Public Health 2021, 14, 638–646. [Google Scholar] [CrossRef] [PubMed]
- Dhouib, W.; Maatoug, J.; Ayouni, I.; Zammit, N.; Ghammem, R.; Fredj, S.B.; Ghannem, H. The incubation period during the pandemic of COVID-19: A systematic review and meta-analysis. Syst. Rev. 2021, 10, 101. [Google Scholar] [CrossRef]
- Jiang, X.; Rayner, S.; Luo, M.-H. Does SARS-CoV-2 has a longer incubation period than SARS and MERS? J. Med. Virol. 2020, 92, 476–478. [Google Scholar] [CrossRef]
- Delamater, P.L.; Street, E.J.; Leslie, T.F.; Yang, Y.T.; Jacobsen, K.H. Complexity of the Basic Reproduction Number (R0). Emerg. Infect. Dis. 2019, 25, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Petersen, E.; Koopmans, M.; Go, U.; Hamer, D.H.; Petrosillo, N.; Castelli, F.; Storgaard, M.; Al Khalili, S.; Simonsen, L. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect. Dis. 2020, 20, e238–e244. [Google Scholar] [CrossRef]
- Liu, Y.; Rocklöv, J. The reproductive number of the Delta variant of SARS-CoV-2 is far higher compared to the ancestral SARS-CoV-2 virus. J. Travel Med. 2021, 28, taab124. [Google Scholar] [CrossRef]
- Liu, Y.; Gayle, A.A.; Wilder-Smith, A.; Rocklöv, J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J. Travel Med. 2020, 27, taaa021. [Google Scholar] [CrossRef] [Green Version]
- Vivanti, A.J.; Vauloup-Fellous, C.; Prevot, S.; Zupan, V.; Suffee, C.; Do Cao, J.; Benachi, A.; De Luca, D. Transplacental transmission of SARS-CoV-2 infection. Nat. Commun. 2020, 11, 3572. [Google Scholar] [CrossRef]
- Mahyuddin, A.P.; Kanneganti, A.; Wong, J.J.L.; Dimri, P.S.; Su, L.L.; Biswas, A.; Illanes, S.E.; Mattar, C.N.Z.; Huang, R.Y.-J.; Choolani, M. Mechanisms and evidence of vertical transmission of infections in pregnancy including SARS-CoV-2s. Prenat. Diagn. 2020, 40, 1655–1670. [Google Scholar] [CrossRef]
- Saadaoui, M.; Kumar, M.; Al Khodor, S. COVID-19 Infection during Pregnancy: Risk of Vertical Transmission, Fetal, and Neonatal Outcomes. J. Pers. Med. 2021, 11, 483. [Google Scholar] [CrossRef]
- Kumar, A.; Prasoon, P.; Kumari, C.; Pareek, V.; Faiq, M.A.; Narayan, R.K.; Kulandhasamy, M.; Kant, K. SARS-CoV-2-specific virulence factors in COVID-19. J. Med. Virol. 2021, 93, 1343–1350. [Google Scholar] [CrossRef]
- Tsatsakis, A.; Petrakis, D.; Nikolouzakis, T.K.; Docea, A.O.; Calina, D.; Vinceti, M.; Goumenou, M.; Kostoff, R.N.; Mamoulakis, C.; Aschner, M.; et al. COVID-19, an opportunity to reevaluate the correlation between long-term effects of anthropogenic pollutants on viral epidemic/pandemic events and prevalence. Food Chem. Toxicol. 2020, 141, 111418. [Google Scholar] [CrossRef]
- Yang, L.; Xie, X.; Tu, Z.; Fu, J.; Xu, D.; Zhou, Y. The signal pathways and treatment of cytokine storm in COVID-19. Signal Transduct. Target. Ther. 2021, 6, 255. [Google Scholar] [CrossRef]
- Chen, R.; Huang, Y.; Quan, J.; Liu, J.; Wang, H.; Billiar, T.R.; Lotze, M.T.; Zeh, H.J.; Kang, R.; Tang, D. HMGB1 as a potential biomarker and therapeutic target for severe COVID-19. Heliyon 2020, 6, e05672. [Google Scholar] [CrossRef]
- Alnajjar, R.; Mostafa, A.; Kandeil, A.; Al-Karmalawy, A.A.J.H. Molecular docking, molecular dynamics, and in vitro studies reveal the potential of angiotensin II receptor blockers to inhibit the COVID-19 main protease. Heliyon 2020, 6, e05641. [Google Scholar] [CrossRef]
- Elmaaty, A.A.; Darwish, K.M.; Khattab, M.; Elhady, S.S.; Salah, M.; Hamed, M.I.A.; Al-Karmalawy, A.A.; Saleh, M.M. In a search for potential drug candidates for combating COVID-19: Computational study revealed salvianolic acid B as a potential therapeutic targeting 3CLpro and spike proteins. J. Biomol. Struct. Dyn. 2021, 39, 1–28. [Google Scholar] [CrossRef]
- Abo Elmaaty, A.; Hamed, M.I.A.; Ismail, M.I.; Elkaeed, E.B.; Abulkhair, H.S.; Khattab, M.; Al-Karmalawy, A.A. Computational Insights on the Potential of Some NSAIDs for Treating COVID-19: Priority Set and Lead Optimization. Molecules 2021, 26, 3772. [Google Scholar] [CrossRef]
- Gheblawi, M.; Wang, K.; Viveiros, A.; Nguyen, Q.; Zhong, J.-C.; Turner, A.J.; Raizada, M.K.; Grant, M.B.; Oudit, G.Y. Angiotensin-Converting Enzyme 2: SARS-CoV-2 Receptor and Regulator of the Renin-Angiotensin System: Celebrating the 20th Anniversary of the Discovery of ACE2. Circ. Res. 2020, 126, 1456–1474. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Murgolo, N.; Therien, A.G.; Howell, B.; Klein, D.; Koeplinger, K.; Lieberman, L.A.; Adam, G.C.; Flynn, J.; McKenna, P.; Swaminathan, G.; et al. SARS-CoV-2 tropism, entry, replication, and propagation: Considerations for drug discovery and development. PLoS Pathog. 2021, 17, e1009225. [Google Scholar] [CrossRef]
- Baruah, C.; Devi, P.; Sharma, D.K. Sequence Analysis and Structure Prediction of SARS-CoV-2 Accessory Proteins 9b and ORF14: Evolutionary Analysis Indicates Close Relatedness to Bat Coronavirus. BioMed Res. Int. 2020, 2020, 7234961. [Google Scholar] [CrossRef]
- Saraste, J.; Prydz, K. Assembly and Cellular Exit of Coronaviruses: Hijacking an Unconventional Secretory Pathway from the Pre-Golgi Intermediate Compartment via the Golgi Ribbon to the Extracellular Space. Cells 2021, 10, 503. [Google Scholar] [CrossRef]
- WHO. Clinical Management of COVID-19: Living Guidance. Available online: https://www.who.int/publications/i/item/WHO-2019-nCoV-clinical-2021-1 (accessed on 19 October 2021).
- Khattab, M.; Al-Karmalawy, A.A. Computational repurposing of benzimidazole anthelmintic drugs as potential colchicine binding site inhibitors. Futur. Med. Chem. 2021, 13, 1623–1638. [Google Scholar] [CrossRef] [PubMed]
- Al-Karmalawy, A.A.; Khattab, M. Molecular modelling of mebendazole polymorphs as a potential colchicine binding site inhibitor. New J. Chem. 2020, 44, 13990–13996. [Google Scholar] [CrossRef]
- Mahmoud, D.B.; Ismail, W.M.; Moatasim, Y.; Kutkat, O.; ElMeshad, A.N.; Ezzat, S.M.; El Deeb, K.S.; El-Fishawy, A.M.; Gomaa, M.R.; Kandeil, A.; et al. Delineating a potent antiviral activity of Cuphea ignea extract loaded nano-formulation against SARS-CoV-2: In silico and in vitro studies. J. Drug Deliv. Sci. Technol. 2021, 66, 102845. [Google Scholar] [CrossRef] [PubMed]
- NIH. Clinical Spectrum of SARS-CoV-2 Infection. Available online: https://www.covid19treatmentguidelines.nih.gov/overview/clinical-spectrum/ (accessed on 19 October 2021).
- Shafran, N.; Shafran, I.; Ben-Zvi, H.; Sofer, S.; Sheena, L.; Krause, I.; Shlomai, A.; Goldberg, E.; Sklan, E.H. Secondary bacterial infection in COVID-19 patients is a stronger predictor for death compared to influenza patients. Sci. Rep. 2021, 11, 12703. [Google Scholar] [CrossRef]
- McFee, R. COVID-19 medical management including World Health Organization (WHO) suggested management strategies. Dis. Mon. 2020, 66, 101068. [Google Scholar] [CrossRef]
- Perez, M.A.; Gordon, A.; Sanchez, F.; Narvaez, F.; Gutierrez, G.; Ortega, O.; Nuñez, A.; Harris, E.; Balmaseda, A. Severe coinfections of dengue and pandemic influenza A H1N1 viruses. Pediatr. Infect. Dis. J. 2010, 29, 1052–1055. [Google Scholar] [CrossRef] [Green Version]
- Drożdżal, S.; Rosik, J.; Lechowicz, K.; Machaj, F.; Szostak, B.; Majewski, P.; Rotter, I.; Kotfis, K. COVID-19: Pain management in patients with SARS-CoV-2 infection—molecular mechanisms, challenges, and perspectives. Brain Sci. 2020, 10, 465. [Google Scholar] [CrossRef]
- Greenhalgh, T.; Koh, G.C.H.; Car, J. Covid-19: A remote assessment in primary care. BMJ 2020, 368, m1182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goossens, H.; Ferech, M.; Vander Stichele, R.; Elseviers, M.; Group, E.P. Outpatient antibiotic use in Europe and association with resistance: A cross-national database study. Lancet 2005, 365, 579–587. [Google Scholar] [CrossRef]
- Llor, C.; Bjerrum, L. Antimicrobial resistance: Risk associated with antibiotic overuse and initiatives to reduce the problem. Ther. Adv. Drug Saf. 2014, 5, 229–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almadhi, M.A.; Abdulrahman, A.; Sharaf, S.A.; AlSaad, D.; Stevenson, N.J.; Atkin, S.L.; AlQahtani, M.M. The high prevalence of asymptomatic SARS-CoV-2 infection reveals the silent spread of COVID-19. Int. J. Infect. Dis. 2021, 105, 656–661. [Google Scholar] [CrossRef]
- Liang, S.T.; Liang, L.T.; Rosen, J.M. COVID-19: A comparison to the 1918 influenza and how we can defeat it. Postgrad. Med. J. 2021, 97, 273. [Google Scholar] [CrossRef] [PubMed]
- Jensen, L.A.; Onyskiw, J.E.; Prasad, N. Meta-analysis of arterial oxygen saturation monitoring by pulse oximetry in adults. Heart Lung 1998, 27, 387–408. [Google Scholar] [CrossRef]
- Zheng, K.I.; Feng, G.; Liu, W.-Y.; Targher, G.; Byrne, C.D.; Zheng, M.-H. Extrapulmonary complications of COVID-19: A multisystem disease? J. Med. Virol. 2021, 93, 323–335. [Google Scholar] [CrossRef]
- Xiong, X.; Chi, J.; Gao, Q. Prevalence and risk factors of thrombotic events on patients with COVID-19: A systematic review and meta-analysis. Thromb. J. 2021, 19, 32. [Google Scholar] [CrossRef]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Poudel, A.; Poudel, Y.; Adhikari, A.; Aryal, B.B.; Dangol, D.; Bajracharya, T.; Maharjan, A.; Gautam, R. D-dimer as a biomarker for assessment of COVID-19 prognosis: D-dimer levels on admission and its role in predicting disease outcome in hospitalized patients with COVID-19. PLoS ONE 2021, 16, e0256744. [Google Scholar] [CrossRef]
- Papali, A.; Adhikari, N.K.; Diaz, J.V.; Dondorp, A.M.; Dünser, M.W.; Jacob, S.T.; Phua, J.; Romain, M.; Schultz, M.J. Infrastructure and organization of adult intensive care units in resource-limited settings. In Sepsis Management in Resource-Limited Settings; Springer: Cham, Switzerland, 2019; pp. 31–68. [Google Scholar] [CrossRef] [Green Version]
- Elebeedy, D.; Elkhatib, W.F.; Kandeil, A.; Ghanem, A.; Kutkat, O.; Alnajjar, R.; Saleh, M.A.; Abd El Maksoud, A.I.; Badawy, I.; Al-Karmalawy, A.A. Anti-SARS-CoV-2 activities of tanshinone IIA, carnosic acid, rosmarinic acid, salvianolic acid, baicalein, and glycyrrhetinic acid between computational and in vitro insights. RSC Adv. 2021, 11, 29267–29286. [Google Scholar] [CrossRef]
- WHO. COVID-19 Weekly Epidemiological Update. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (accessed on 27 September 2021).
- Soltan, M.A.; Elbassiouny, N.; Gamal, H.; Elkaeed, E.B.; Eid, R.A.; Eldeen, M.A.; Al-Karmalawy, A.A. In Silico Prediction of a Multitope Vaccine against Moraxella catarrhalis: Reverse Vaccinology and Immunoinformatics. Vaccines 2021, 9, 669. [Google Scholar] [CrossRef] [PubMed]
- Soltan, M.A.; Eldeen, M.A.; Elbassiouny, N.; Mohamed, I.; El-damasy, D.A.; Fayad, E.; Abu Ali, O.A.; Raafat, N.; Eid, R.A.; Al-Karmalawy, A.A. Proteome Based Approach Defines Candidates for Designing a Multitope Vaccine against the Nipah Virus. Int. J. Mol. Sci. 2021, 22, 9330. [Google Scholar] [CrossRef] [PubMed]
- Shehata, M.M.; Mahmoud, S.H.; Tarek, M.; Al-Karmalawy, A.A.; Mahmoud, A.; Mostafa, A.; Elhefnawi, M.M.; Ali, M.A. In Silico and In Vivo Evaluation of SARS-CoV-2 Predicted Epitopes-Based Candidate Vaccine. Molecules 2021, 26, 6182. [Google Scholar] [CrossRef]
- Yang, J.; Petitjean, S.J.L.; Koehler, M.; Zhang, Q.; Dumitru, A.C.; Chen, W.; Derclaye, S.; Vincent, S.P.; Soumillion, P.; Alsteens, D. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun. 2020, 11, 4541. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, C.; Xu, X.-f.; Xu, W.; Liu, S.-w. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef] [PubMed]
- Boson, B.; Legros, V.; Zhou, B.; Siret, E.; Mathieu, C.; Cosset, F.L.; Lavillette, D.; Denolly, S. The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus-like particles. J. Biol. Chem. 2021, 296, 100111. [Google Scholar] [CrossRef] [PubMed]
- Cubuk, J.; Alston, J.J.; Incicco, J.J.; Singh, S.; Stuchell-Brereton, M.D.; Ward, M.D.; Zimmerman, M.I.; Vithani, N.; Griffith, D.; Wagoner, J.A.; et al. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat. Commun. 2021, 12, 1936. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.; Chaudhary, J.K.; Jain, N.; Chaudhary, P.K.; Khanra, S.; Dhamija, P.; Sharma, A.; Kumar, A.; Handu, S. Role of Structural and Non-Structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19. Cells 2021, 10, 821. [Google Scholar] [CrossRef]
- Mariano, G.; Farthing, R.J.; Lale-Farjat, S.L.M.; Bergeron, J.R.C. Structural Characterization of SARS-CoV-2: Where We Are, and Where We Need to Be. Front. Mol. Biosci. 2020, 7, 344. [Google Scholar] [CrossRef]
- Khan, W.H.; Hashmi, Z.; Goel, A.; Ahmad, R.; Gupta, K.; Khan, N.; Alam, I.; Ahmed, F.; Ansari, M.A. COVID-19 Pandemic and Vaccines Update on Challenges and Resolutions. Front. Cell. Infect. Microbiol. 2021, 11, 690621. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F. SARS-CoV-2 vaccines in development. Nature 2020, 586, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Shehata, M.M.; Mostafa, A.; Teubner, L.; Mahmoud, S.H.; Kandeil, A.; Elshesheny, R.; Frantz, R.; La Pietra, L.; Pleschka, S.; Osman, A.; et al. Bacterial Outer Membrane Vesicles (OMVs)-based Dual Vaccine for Influenza A H1N1 Virus and MERS-CoV. Vaccines 2019, 7, 46. [Google Scholar] [CrossRef] [Green Version]
- Rappazzo, C.G.; Watkins, H.C.; Guarino, C.M.; Chau, A.; Lopez, J.L.; DeLisa, M.P.; Leifer, C.A.; Whittaker, G.R.; Putnam, D. Recombinant M2e outer membrane vesicle vaccines protect against lethal influenza A challenge in BALB/c mice. Vaccine 2016, 34, 1252–1258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaspar, E.B.; Prudencio, C.R.; De Gaspari, E. Experimental studies using OMV in a new platform of SARS-CoV-2 vaccines. Hum. Vaccines Immunother. 2021, 17, 2965–2968. [Google Scholar] [CrossRef]
- Riad, A.; Hocková, B.; Kantorová, L.; Slávik, R.; Spurná, L.; Stebel, A.; Havriľak, M.; Klugar, M. Side Effects of mRNA-Based COVID-19 Vaccine: Nationwide Phase IV Study among Healthcare Workers in Slovakia. Pharmaceuticals 2021, 14, 873. [Google Scholar] [CrossRef]
- Doerfler, W. Adenoviral Vector DNA-and SARS-CoV-2 mRNA-Based Covid-19 Vaccines: Possible Integration into the Human Genome—Are Adenoviral Genes Expressed in Vector-based Vaccines? Virus Res. 2021, 302, 198466. [Google Scholar] [CrossRef]
- Muir, K.-L.; Kallam, A.; Koepsell, S.A.; Gundabolu, K. Thrombotic Thrombocytopenia after Ad26.COV2.S Vaccination. N. Engl. J. Med. 2021, 384, 1964–1965. [Google Scholar] [CrossRef]
- Meo, S.A.; Bukhari, I.A.; Akram, J.; Meo, A.S.; Klonoff, D.C. COVID-19 vaccines: Comparison of biological, pharmacological characteristics and adverse effects of Pfizer/BioNTech and Moderna Vaccines. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 1663–1669. [Google Scholar] [CrossRef]
- Bozkurt, B.; Kamat, I.; Hotez, P.J. Myocarditis With COVID-19 mRNA Vaccines. Circulation 2021, 144, 471–484. [Google Scholar] [CrossRef]
- Verma, A.K.; Lavine, K.J.; Lin, C.-Y. Myocarditis after Covid-19 mRNA Vaccination. N. Engl. J. Med. 2021, 385, 1332–1334. [Google Scholar] [CrossRef] [PubMed]
- Barda, N.; Dagan, N.; Ben-Shlomo, Y.; Kepten, E.; Waxman, J.; Ohana, R.; Hernan, M.A.; Lipsitch, M.; Kohane, I.; Netzer, D.; et al. Safety of the BNT162b2 mRNA Covid-19 Vaccine in a Nationwide Setting. N. Engl. J. Med. 2021, 385, 1078–1090. [Google Scholar] [CrossRef] [PubMed]
- Witberg, G.; Barda, N.; Hoss, S.; Richter, I.; Wiessman, M.; Aviv, Y.; Grinberg, T.; Auster, O.; Dagan, N.; Balicer, R.D.; et al. Myocarditis after Covid-19 Vaccination in a Large Health Care Organization. N. Engl. J. Med. 2021. [Google Scholar] [CrossRef]
- Greinacher, A.; Thiele, T.; Warkentin, T.E.; Weisser, K.; Kyrle, P.A.; Eichinger, S. Thrombotic Thrombocytopenia after ChAdOx1 nCov-19 Vaccination. N. Engl. J. Med. 2021, 384, 2092–2101. [Google Scholar] [CrossRef] [PubMed]
- Simpson, C.R.; Shi, T.; Vasileiou, E.; Katikireddi, S.V.; Kerr, S.; Moore, E.; McCowan, C.; Agrawal, U.; Shah, S.A.; Ritchie, L.D.; et al. First-dose ChAdOx1 and BNT162b2 COVID-19 vaccines and thrombocytopenic, thromboembolic and hemorrhagic events in Scotland. Nat. Med. 2021, 27, 1290–1297. [Google Scholar] [CrossRef]
- EMA. COVID-19 Vaccine AstraZeneca: Benefits still Outweigh the Risks Despite Possible Link to Rare Blood Clots with Low Blood Platelets. Available online: https://www.ema.europa.eu/en/news/covid-19-vaccine-astrazeneca-benefits-still-outweigh-risks-despite-possible-link-rare-blood-clots (accessed on 26 October 2021).
- Poland, G.A.; Ovsyannikova, I.G.; Kennedy, R.B. SARS-CoV-2 immunity: Review and applications to phase 3 vaccine candidates. Lancet 2020, 396, 1595–1606. [Google Scholar] [CrossRef]
- Tregoning, J.S.; Flight, K.E.; Higham, S.L.; Wang, Z.; Pierce, B.F. Progress of the COVID-19 vaccine effort: Viruses, vaccines and variants versus efficacy, effectiveness and escape. Nat. Rev. Immunol. 2021, 21, 626–636. [Google Scholar] [CrossRef] [PubMed]
- Sa Ribero, M.; Jouvenet, N.; Dreux, M.; Nisole, S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020, 16, e1008737. [Google Scholar] [CrossRef]
- Oh, S.J.; Shin, O.S. SARS-CoV-2 Nucleocapsid Protein Targets RIG-I-Like Receptor Pathways to Inhibit the Induction of Interferon Response. Cells 2021, 10, 530. [Google Scholar] [CrossRef]
- Coperchini, F.; Chiovato, L.; Croce, L.; Magri, F.; Rotondi, M. The cytokine storm in COVID-19: An overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020, 53, 25–32. [Google Scholar] [CrossRef]
- Costela-Ruiz, V.J.; Illescas-Montes, R.; Puerta-Puerta, J.M.; Ruiz, C.; Melguizo-Rodríguez, L. SARS-CoV-2 infection: The role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020, 54, 62–75. [Google Scholar] [CrossRef]
- Fara, A.; Mitrev, Z.; Rosalia, R.A.; Assas, B.M. Cytokine storm and COVID-19: A chronicle of pro-inflammatory cytokines. Open Biol. 2020, 10, 200160. [Google Scholar] [CrossRef]
- Satarker, S.; Tom, A.A.; Shaji, R.A.; Alosious, A.; Luvis, M.; Nampoothiri, M. JAK-STAT Pathway Inhibition and their Implications in COVID-19 Therapy. Postgrad. Med. 2021, 133, 489–507. [Google Scholar] [CrossRef]
- Van Eeden, C.; Khan, L.; Osman, M.S.; Cohen Tervaert, J.W. Natural Killer Cell Dysfunction and Its Role in COVID-19. Int. J. Mol. Sci. 2020, 21, 6351. [Google Scholar] [CrossRef]
- Wu, J.; Liang, B.; Chen, C.; Wang, H.; Fang, Y.; Shen, S.; Yang, X.; Wang, B.; Chen, L.; Chen, Q.; et al. SARS-CoV-2 infection induces sustained humoral immune responses in convalescent patients following symptomatic COVID-19. Nat. Commun. 2021, 12, 1813. [Google Scholar] [CrossRef]
- Gaebler, C.; Wang, Z.; Lorenzi, J.C.C.; Muecksch, F.; Finkin, S.; Tokuyama, M.; Cho, A.; Jankovic, M.; Schaefer-Babajew, D.; Oliveira, T.Y.; et al. Evolution of antibody immunity to SARS-CoV-2. Nature 2021, 591, 639–644. [Google Scholar] [CrossRef]
- Liu, W.J.; Zhao, M.; Liu, K.; Xu, K.; Wong, G.; Tan, W.; Gao, G.F. T-cell immunity of SARS-CoV: Implications for vaccine development against MERS-CoV. Antivir. Res. 2017, 137, 82–92. [Google Scholar] [CrossRef]
- Xiao, K.; Yang, H.; Liu, B.; Pang, X.; Du, J.; Liu, M.; Liu, Y.; Jing, X.; Chen, J.; Deng, S.; et al. Antibodies Can Last for More Than 1 Year After SARS-CoV-2 Infection: A Follow-Up Study From Survivors of COVID-19. Front. Med. 2021, 8, 3. [Google Scholar] [CrossRef]
- Zeng, F.; Wu, M.; Wang, J.; Li, J.; Hu, G.; Wang, L. Over 1-year duration and age difference of SARS-CoV-2 antibodies in convalescent COVID-19 patients. J. Med. Virol. 2021, 93, 6506–6511. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yu, D.; Wu, X.; Liang, H.; Zhou, Z.; Xie, Y.; Li, T.; Wu, J.; Lu, F.; Feng, L.; et al. Twelve-month specific IgG response to SARS-CoV-2 receptor-binding domain among COVID-19 convalescent plasma donors in Wuhan. Nat. Commun. 2021, 12, 4144. [Google Scholar] [CrossRef] [PubMed]
- Glück, V.; Grobecker, S.; Tydykov, L.; Salzberger, B.; Glück, T.; Weidlich, T.; Bertok, M.; Gottwald, C.; Wenzel, J.J.; Gessner, A.; et al. SARS-CoV-2-directed antibodies persist for more than six months in a cohort with mild to moderate COVID-19. Infection 2021, 49, 739–746. [Google Scholar] [CrossRef]
- Dobaño, C.; Ramírez-Morros, A.; Alonso, S.; Vidal-Alaball, J.; Ruiz-Olalla, G.; Vidal, M.; Rubio, R.; Cascant, E.; Parras, D.; Rodrigo Melero, N.; et al. Persistence and baseline determinants of seropositivity and reinfection rates in health care workers up to 12.5 months after COVID-19. BMC Med. 2021, 19, 155. [Google Scholar] [CrossRef] [PubMed]
- Harrington, W.E.; Trakhimets, O.; Andrade, D.V.; Dambrauskas, N.; Raappana, A.; Jiang, Y.; Houck, J.; Selman, W.; Yang, A.; Vigdorovich, V.; et al. Rapid decline of neutralizing antibodies is associated with decay of IgM in adults recovered from mild COVID-19. Cell Rep. Med. 2021, 2, 100253. [Google Scholar] [CrossRef]
- Jung, J.H.; Rha, M.-S.; Sa, M.; Choi, H.K.; Jeon, J.H.; Seok, H.; Park, D.W.; Park, S.-H.; Jeong, H.W.; Choi, W.S.; et al. SARS-CoV-2-specific T cell memory is sustained in COVID-19 convalescent patients for 10 months with successful development of stem cell-like memory T cells. Nat. Commun. 2021, 12, 4043. [Google Scholar] [CrossRef]
- Zotta, A.; Hooftman, A.; O’Neill, L.A.J. SARS-CoV-2 targets MAVS for immune evasion. Nat. Cell Biol. 2021, 23, 682–683. [Google Scholar] [CrossRef] [PubMed]
- Kasuga, Y.; Zhu, B.; Jang, K.-J.; Yoo, J.-S. Innate immune sensing of coronavirus and viral evasion strategies. Exp. Mol. Med. 2021, 53, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Beyer, D.K.; Forero, A. Mechanisms of Antiviral Immune Evasion of SARS-CoV-2. J. Mol. Biol. 2021, 167265. [Google Scholar] [CrossRef]
- Klein, S.; Cortese, M.; Winter, S.L.; Wachsmuth-Melm, M.; Neufeldt, C.J.; Cerikan, B.; Stanifer, M.L.; Boulant, S.; Bartenschlager, R.; Chlanda, P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020, 11, 5885. [Google Scholar] [CrossRef]
- Yuan, S.; Balaji, S.; Lomakin, I.B.; Xiong, Y. Coronavirus Nsp1: Immune Response Suppression and Protein Expression Inhibition. Front. Microbiol. 2021, 12, 752214. [Google Scholar] [CrossRef]
- Moustaqil, M.; Ollivier, E.; Chiu, H.P.; Van Tol, S.; Rudolffi-Soto, P.; Stevens, C.; Bhumkar, A.; Hunter, D.J.B.; Freiberg, A.N.; Jacques, D.; et al. SARS-CoV-2 proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): Implications for disease presentation across species. Emerg. Microbes Infect. 2021, 10, 178–195. [Google Scholar] [CrossRef]
- Miorin, L.; Kehrer, T.; Sanchez-Aparicio, M.T.; Zhang, K.; Cohen, P.; Patel, R.S.; Cupic, A.; Makio, T.; Mei, M.; Moreno, E.; et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 28344–28354. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, Y.; Li, Y.; Huang, F.; Luo, B.; Yuan, Y.; Xia, B.; Ma, X.; Yang, T.; Yu, F.; et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Ι. Proc. Natl. Acad. Sci. USA 2021, 118, e2024202118. [Google Scholar] [CrossRef]
- Yuen, C.K.; Lam, J.Y.; Wong, W.M.; Mak, L.F.; Wang, X.; Chu, H.; Cai, J.P.; Jin, D.Y.; To, K.K.; Chan, J.F.; et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microbes Infect. 2020, 9, 1418–1428. [Google Scholar] [CrossRef]
- Rehwinkel, J.; Gack, M.U. RIG-I-like receptors: Their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020, 20, 537–551. [Google Scholar] [CrossRef]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Pettoello-Mantovani, M.; Carrasco-Sanz, A.; Huss, G.; Mestrovic, J.; Vural, M.; Pop, T.L.; Ferrara, P.; Somekh, E.; Mujkic, A.; Hoey, H.; et al. Viewpoint of the European Pediatric Societies over Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Vaccination in Children Younger Than Age 12 Years Amid Return to School and the Surging Virus Variants. J. Pediatr. 2021, in press. [Google Scholar] [CrossRef]
- Oude Munnink, B.B.; Worp, N.; Nieuwenhuijse, D.F.; Sikkema, R.S.; Haagmans, B.; Fouchier, R.A.M.; Koopmans, M. The next phase of SARS-CoV-2 surveillance: Real-time molecular epidemiology. Nat. Med. 2021, 27, 1518–1524. [Google Scholar] [CrossRef] [PubMed]
- Tegally, H.; Wilkinson, E.; Lessells, R.J.; Giandhari, J.; Pillay, S.; Msomi, N.; Mlisana, K.; Bhiman, J.N.; von Gottberg, A.; Walaza, S.; et al. Sixteen novel lineages of SARS-CoV-2 in South Africa. Nat. Med. 2021, 27, 440–446. [Google Scholar] [CrossRef]
- Wilkinson, E.; Giovanetti, M.; Tegally, H.; San, J.E.; Lessells, R.; Cuadros, D.; Martin, D.P.; Rasmussen, D.A.; Zekri, A.-R.N.; Sangare, A.K.; et al. A year of genomic surveillance reveals how the SARS-CoV-2 pandemic unfolded in Africa. Science 2021, 374, eabj4336. [Google Scholar] [CrossRef] [PubMed]
- Lopez Bernal, J.; Andrews, N.; Gower, C.; Gallagher, E.; Simmons, R.; Thelwall, S.; Stowe, J.; Tessier, E.; Groves, N.; Dabrera, G.; et al. Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant. N. Engl. J. Med. 2021, 385, 585–594. [Google Scholar] [CrossRef]
- Ding, C.; He, J.; Zhang, X.; Jiang, C.; Sun, Y.; Zhang, Y.; Chen, Q.; He, H.; Li, W.; Xie, J.; et al. Crucial Mutations of Spike Protein on SARS-CoV-2 Evolved to Variant Strains Escaping Neutralization of Convalescent Plasmas and RBD-Specific Monoclonal Antibodies. Front. Immunol. 2021, 12, 3231. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Steinkellner, G.; Köchl, K.; Gruber, K.; Gruber, C.C. Serine 477 plays a crucial role in the interaction of the SARS-CoV-2 spike protein with the human receptor ACE2. Sci. Rep. 2021, 11, 4320. [Google Scholar] [CrossRef]
- Chen, J.; Gao, K.; Wang, R.; Wei, G.-W. Revealing the threat of emerging SARS-CoV-2 mutations to antibody therapies. bioRxiv 2021. [Google Scholar] [CrossRef]
- Castelli, M.; Baj, A.; Criscuolo, E.; Ferrarese, R.; Diotti, R.A.; Sampaolo, M.; Novazzi, F.; Dalla Gasperina, D.; Focosi, D.; Ferrari, D.; et al. Characterization of a Lineage C.36 SARS-CoV-2 Isolate with Reduced Susceptibility to Neutralization Circulating in Lombardy, Italy. Viruses 2021, 13, 1514. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Garcia-Knight, M.A.; Khalid, M.M.; Servellita, V.; Wang, C.; Morris, M.K.; Sotomayor-González, A.; Glasner, D.R.; Reyes, K.R.; Gliwa, A.S.; et al. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell 2021, 184, 3426–3437.e8. [Google Scholar] [CrossRef] [PubMed]
- McCallum, M.; Bassi, J.; De Marco, A.; Chen, A.; Walls, A.C.; Di Iulio, J.; Tortorici, M.A.; Navarro, M.J.; Silacci-Fregni, C.; Saliba, C.; et al. SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science 2021, 373, 648–654. [Google Scholar] [CrossRef] [PubMed]
- Planas, D.; Veyer, D.; Baidaliuk, A.; Staropoli, I.; Guivel-Benhassine, F.; Rajah, M.M.; Planchais, C.; Porrot, F.; Robillard, N.; Puech, J.; et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 2021, 596, 276–280. [Google Scholar] [CrossRef] [PubMed]
- Vanhove, B.; Marot, S.; So, R.T.; Gaborit, B.; Evanno, G.; Malet, I.; Lafrogne, G.; Mevel, E.; Ciron, C.; Royer, P.-J.; et al. XAV-19, a swine glyco-humanized polyclonal antibody against SARS-CoV-2 Spike receptor-binding domain, targets multiple epitopes and broadly neutralizes variants. bioRxiv 2021. [Google Scholar] [CrossRef]
- Khattab, M.; Al-Karmalawy, A.A. Revisiting Activity of Some Nocodazole Analogues as a Potential Anticancer Drugs Using Molecular Docking and DFT Calculations. Front. Chem. 2021, 9, 92. [Google Scholar] [CrossRef]
- Samra, R.M.; Soliman, A.F.; Zaki, A.A.; Ashour, A.; Al-Karmalawy, A.A.; Hassan, M.A.; Zaghloul, A.M. Bioassay-guided isolation of a new cytotoxic ceramide from Cyperus rotundus L. S. Afr. J. Bot. 2021, 139, 210–216. [Google Scholar] [CrossRef]
- Eliaa, S.G.; Al-Karmalawy, A.A.; Saleh, R.M.; Elshal, M.F. Empagliflozin and Doxorubicin Synergistically Inhibit the Survival of Triple-Negative Breast Cancer Cells via Interfering with the mTOR Pathway and Inhibition of Calmodulin: In Vitro and Molecular Docking Studies. ACS Pharmacol. Transl. Sci. 2020, 3, 1330–1338. [Google Scholar] [CrossRef]
- Al-Karmalawy, A.A.; Elshal, M.F. Concanavalin-A shows synergistic cytotoxicity with tamoxifen via inducing apoptosis in estrogen receptor-positive breast cancer: In vitro and molecular docking studies. Pharm. Sci. 2021. [Google Scholar] [CrossRef]
- Ghanem, A.; Emara, H.A.; Muawia, S.; Abd El Maksoud, A.I.; Al-Karmalawy, A.A.; Elshal, M.F. Tanshinone IIA synergistically enhances the antitumor activity of doxorubicin by interfering with the PI3K/AKT/mTOR pathway and inhibition of topoisomerase II: In vitro and molecular docking studies. New J. Chem. 2020, 44, 17374–17381. [Google Scholar] [CrossRef]
- Zaki, A.A.; Al-Karmalawy, A.A.; Khodir, A.E.; El-Amier, Y.A.; Ashour, A. Isolation of cytotoxic active compounds from Reichardia tingitana with investigation of apoptosis mechanistic induction: In silico, in vitro, and SAR studies. S. Afr. J. Bot. 2021, 144, 115–123. [Google Scholar] [CrossRef]
- Zaki, A.A.; Ashour, A.; Elhady, S.S.; Darwish, K.M.; Al-Karmalawy, A.A. Calendulaglycoside A Showing Potential Activity Against SARS-CoV-2 Main Protease: Molecular Docking, Molecular Dynamics, and SAR Studies. J. Tradit. Complement. Med. 2021, in press. [Google Scholar] [CrossRef] [PubMed]
- El-Demerdash, A.; Al-Karmalawy, A.A.; Abdel-Aziz, T.M.; Elhady, S.S.; Darwish, K.M.; Hassan, A.H.E. Investigating the structure–activity relationship of marine natural polyketides as promising SARS-CoV-2 main protease inhibitors. RSC Adv. 2021, 11, 31339–31363. [Google Scholar] [CrossRef]
- Abdallah, A.E.; Alesawy, M.S.; Eissa, S.I.; El-Fakharany, E.M.; Kalaba, M.H.; Sharaf, M.H.; Abo Shama, N.M.; Mahmoud, S.H.; Mostafa, A.; Al-Karmalawy, A.A.; et al. Design and synthesis of new 4-(2-nitrophenoxy)benzamide derivatives as potential antiviral agents: Molecular modeling and in vitro antiviral screening. New J. Chem. 2021, 45, 16557–16571. [Google Scholar] [CrossRef]
- Al-Karmalawy, A.A.; Dahab, M.A.; Metwaly, A.M.; Elhady, S.S.; Elkaeed, E.B.; Eissa, I.H.; Darwish, K.M. Molecular Docking and Dynamics Simulation Revealed the Potential Inhibitory Activity of ACEIs Against SARS-CoV-2 Targeting the hACE2 Receptor. Front. Chem. 2021, 9, 21. [Google Scholar] [CrossRef]
- Budweiser, S.; Baş, Ş.; Jörres, R.A.; Engelhardt, S.; von Delius, S.; Lenherr, K.; Deerberg-Wittram, J.; Bauer, A. Patients’ treatment limitations as predictive factor for mortality in COVID-19: Results from hospitalized patients of a hotspot region for SARS-CoV-2 infections. Respir. Res. 2021, 22, 168. [Google Scholar] [CrossRef] [PubMed]
- Williamson, E.J.; Walker, A.J.; Bhaskaran, K.; Bacon, S.; Bates, C.; Morton, C.E.; Curtis, H.J.; Mehrkar, A.; Evans, D.; Inglesby, P.; et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature 2020, 584, 430–436. [Google Scholar] [CrossRef]
- Salter, A.; Fox, R.J.; Newsome, S.D.; Halper, J.; Li, D.K.B.; Kanellis, P.; Costello, K.; Bebo, B.; Rammohan, K.; Cutter, G.R.; et al. Outcomes and Risk Factors Associated With SARS-CoV-2 Infection in a North American Registry of Patients With Multiple Sclerosis. JAMA Neurol. 2021, 78, 699–708. [Google Scholar] [CrossRef] [PubMed]
- Furst, D.E. Pharmacokinetics of hydroxychloroquine and chloroquine during treatment of rheumatic diseases. Lupus 1996, 5, 11–15. [Google Scholar] [CrossRef]
- Lim, H.-S.; Im, J.-S.; Cho, J.-Y.; Bae, K.-S.; Klein, T.A.; Yeom, J.-S.; Kim, T.-S.; Choi, J.-S.; Jang, I.-J.; Park, J.-W. Pharmacokinetics of hydroxychloroquine and its clinical implications in chemoprophylaxis against malaria caused by Plasmodium vivax. Antimicrob. Agents Chemother. 2009, 53, 1468–1475. [Google Scholar] [CrossRef] [Green Version]
- Devaux, C.A.; Rolain, J.-M.; Colson, P.; Raoult, D. New insights on the antiviral effects of chloroquine against coronavirus: What to expect for COVID-19? Int. J. Antimicrob. Agents 2020, 55, 105938. [Google Scholar] [CrossRef] [PubMed]
- Esper, R.B.; da Silva, R.S.; Oikawa, F.T.C.; Castro, M.M.; Razuk-Filho, A.; Batista, P.; Lotze, S.; da Rocha, C.N.; de Sá Cunha Filho, R.; de Oliveira, S. Empirical Treatment with Hydroxychloroquine and Azithromycin for Suspected Cases of COVID-19 Followed-Up by Telemedicine. Available online: http://blog.couvelard.com/herve/files/etude_bresil.pdf (accessed on 10 October 2021).
- Sarhan, A.A.; Ashour, N.A.; Al-Karmalawy, A.A. The journey of antimalarial drugs against SARS-CoV-2: Review article. Inform. Med. Unlocked 2021, 24, 100604. [Google Scholar] [CrossRef]
- Boulware, D.R.; Pullen, M.F.; Bangdiwala, A.S.; Pastick, K.A.; Lofgren, S.M.; Okafor, E.C.; Skipper, C.P.; Nascene, A.A.; Nicol, M.R.; Abassi, M.; et al. A Randomized Trial of Hydroxychloroquine as Postexposure Prophylaxis for Covid-19. N. Engl. J. Med. 2020, 383, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Axfors, C.; Schmitt, A.M.; Janiaud, P.; van’t Hooft, J.; Abd-Elsalam, S.; Abdo, E.F.; Abella, B.S.; Akram, J.; Amaravadi, R.K.; Angus, D.C.; et al. Mortality outcomes with hydroxychloroquine and chloroquine in COVID-19 from an international collaborative meta-analysis of randomized trials. Nat. Commun. 2021, 12, 2349. [Google Scholar] [CrossRef]
- Self, W.H.; Semler, M.W.; Leither, L.M.; Casey, J.D.; Angus, D.C.; Brower, R.G.; Chang, S.Y.; Collins, S.P.; Eppensteiner, J.C.; Filbin, M.R.; et al. Effect of Hydroxychloroquine on Clinical Status at 14 Days in Hospitalized Patients With COVID-19: A Randomized Clinical Trial. JAMA 2020, 324, 2165–2176. [Google Scholar] [CrossRef]
- Bryant, A.; Lawrie, T.A.; Dowswell, T.; Fordham, E.J.; Scott, M.; Hill, S.R.; Tham, T.C. Ivermectin for prevention and treatment of COVID-19 infection: A systematic review, meta-analysis and trial sequential analysis to inform clinical guidelines. Am. J. Ther. 2021, 28. [Google Scholar] [CrossRef] [PubMed]
- Sharun, K.; Dhama, K.; Patel, S.K.; Pathak, M.; Tiwari, R.; Singh, B.R.; Sah, R.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J.; Leblebicioglu, H. Ivermectin, a new candidate therapeutic against SARS-CoV-2/COVID-19. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 23. [Google Scholar] [CrossRef] [PubMed]
- Caly, L.; Druce, J.D.; Catton, M.G.; Jans, D.A.; Wagstaff, K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020, 178, 104787. [Google Scholar] [CrossRef]
- López-Medina, E.; López, P.; Hurtado, I.C.; Dávalos, D.M.; Ramirez, O.; Martínez, E.; Díazgranados, J.A.; Oñate, J.M.; Chavarriaga, H.; Herrera, S. Effect of ivermectin on time to resolution of symptoms among adults with mild COVID-19: A randomized clinical trial. JAMA 2021, 325, 1426–1435. [Google Scholar] [CrossRef]
- Pott-Junior, H.; Paoliello, M.M.B.; Miguel, A.d.Q.C.; da Cunha, A.F.; de Melo Freire, C.C.; Neves, F.F.; de Avó, L.R.d.S.; Roscani, M.G.; Dos Santos, S.D.S.; Chachá, S.G.F. Use of ivermectin in the treatment of Covid-19: A pilot trial. Toxicol. Rep. 2021, 8, 505–510. [Google Scholar] [CrossRef]
- Mahmoud, D.B.; Shitu, Z.; Mostafa, A. Drug repurposing of nitazoxanide: Can it be an effective therapy for COVID-19? J. Genet. Eng. Biotechnol. 2020, 18, 35. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, A.; Kandeil, A.; Elshaier, Y.A.M.M.; Kutkat, O.; Moatasim, Y.; Rashad, A.A.; Shehata, M.; Gomaa, M.R.; Mahrous, N.; Mahmoud, S.H.; et al. FDA-Approved Drugs with Potent In Vitro Antiviral Activity against Severe Acute Respiratory Syndrome Coronavirus 2. Pharmaceuticals 2020, 13, 443. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, A.J.; Jahan, S.; Ashraf, S.A.; Alreshidi, M.; Ashraf, M.S.; Patel, M.; Snoussi, M.; Singh, R.; Adnan, M. Current status and strategic possibilities on potential use of combinational drug therapy against COVID-19 caused by SARS-CoV-2. J. Biomol. Struct. Dyn. 2020, 39, 1–14. [Google Scholar] [CrossRef]
- Khan, Z.; Ghafoor, D.; Khan, A.; Ualiyeva, D.; Khan, S.A.; Bilal, H.; Khan, B.; Khan, A.; Sajjad, W. Diagnostic approaches and potential therapeutic options for coronavirus disease (COVID-19). New Microbes New Infect. 2020, 38, 100770. [Google Scholar] [CrossRef]
- Blum, V.F.; Cimerman, S.; Hunter, J.R.; Tierno, P.; Lacerda, A.; Soeiro, A.; Cardoso, F.; Bellei, N.C.; Maricato, J.; Mantovani, N. Nitazoxanide superiority to placebo to treat moderate COVID-19–A Pilot prove of concept randomized double-blind clinical trial. EClinicalMedicine 2021, 37, 100981. [Google Scholar] [CrossRef]
- Braga, L.; Ali, H.; Secco, I.; Chiavacci, E.; Neves, G.; Goldhill, D.; Penn, R.; Jimenez-Guardeño, J.M.; Ortega-Prieto, A.M.; Bussani, R.; et al. Drugs that inhibit TMEM16 proteins block SARS-CoV-2 spike-induced syncytia. Nature 2021, 594, 88–93. [Google Scholar] [CrossRef]
- Xu, J.; Shi, P.-Y.; Li, H.; Zhou, J. Broad Spectrum Antiviral Agent Niclosamide and Its Therapeutic Potential. ACS Infect. Dis. 2020, 6, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Yousef, A. The Role of Autophagy in Anti-Wolbachia Antibiotic Therapy. Ph.D. Thesis, Liverpool School of Tropical Medicine, University of Liverpool, Liverpool, UK, 2019. [Google Scholar]
- Kunnumakkara, A.B.; Rana, V.; Parama, D.; Banik, K.; Girisa, S.; Sahu, H.; Thakur, K.K.; Dutta, U.; Garodia, P.; Gupta, S.C. COVID-19, cytokines, inflammation, and spices: How are they related? Life Sci. 2021, 248, 119201. [Google Scholar] [CrossRef]
- Mazzon, M.; Ortega-Prieto, A.M.; Imrie, D.; Luft, C.; Hess, L.; Czieso, S.; Grove, J.; Skelton, J.K.; Farleigh, L.; Bugert, J.J. Identification of broad-spectrum antiviral compounds by targeting viral entry. Viruses 2019, 11, 176. [Google Scholar] [CrossRef]
- Backer, V.; Sjöbring, U.; Sonne, J.; Weiss, A.; Hostrup, M.; Johansen, H.K.; Becker, V.; Sonne, D.P.; Balchen, T.; Jellingsø, M. A randomized, double-blind, placebo-controlled phase 1 trial of inhaled and intranasal niclosamide: A broad spectrum antiviral candidate for treatment of COVID-19. Lancet Reg. Health-Eur. 2021, 4, 100084. [Google Scholar] [CrossRef]
- Abdulamir, A.S.; Gorial, F.I.; Saaedi, S.J.; Maulood, M.F.; Hashim, H.A. Effectiveness and Safety of Niclosamaide as Add-on Therapy to the Standard of Care Measures in COVID-19 Management: Randomized controlled clinical trial. medRxiv 2021. [Google Scholar] [CrossRef]
- Khwaza, V.; Mbese, Z.; Aderibigbe, B.A.; Oyedeji, O.O. Therapeutic Efficacy of Antibiotics in the Treatment of Chronic Diseases. In Antibiotic Materials in Healthcare; Elsevier: Amsterdam, The Netherlands, 2020; pp. 11–32. [Google Scholar]
- Shuchi, N.S. A Study Comparing the Prescribing Pattern of Antibiotics for Fever between Dhaka City and Different Villages in Bangladesh. Ph.D. Thesis, Brac University, Dhaka, Bangladesh, 2019. [Google Scholar]
- Hinks, T.S.C.; Cureton, L.; Knight, R.; Wang, A.; Cane, J.L.; Barber, V.S.; Black, J.; Dutton, S.J.; Melhorn, J.; Jabeen, M.; et al. Azithromycin versus standard care in patients with mild-to-moderate COVID-19 (ATOMIC2): An open-label, randomised trial. Lancet Respir. Med. 2021, 9, 1130–1140. [Google Scholar] [CrossRef]
- Group, P.T.C. Azithromycin for community treatment of suspected COVID-19 in people at increased risk of an adverse clinical course in the UK (PRINCIPLE): A randomised, controlled, open-label, adaptive platform trial. Lancet 2021, 397, 1063–1074. [Google Scholar] [CrossRef]
- Oldenburg, C.E.; Pinsky, B.A.; Brogdon, J.; Chen, C.; Ruder, K.; Zhong, L.; Nyatigo, F.; Cook, C.A.; Hinterwirth, A.; Lebas, E.; et al. Effect of Oral Azithromycin vs Placebo on COVID-19 Symptoms in Outpatients With SARS-CoV-2 Infection: A Randomized Clinical Trial. JAMA 2021, 326, 490–498. [Google Scholar] [CrossRef]
- Fitton, A. The quinolones. Clin. Pharmacokinet. 1992, 22, 1–11. [Google Scholar] [CrossRef]
- Wolf, J.S.; Bennett, C.J.; Dmochowski, R.R.; Hollenbeck, B.K.; Pearle, M.S.; Schaeffer, A.J. Best practice policy statement on urologic surgery antimicrobial prophylaxis. J. Urol. 2008, 179, 1379–1390. [Google Scholar] [CrossRef]
- Xi, W.-N.; Jin, D.; Sun, K.; Yu, R.-Y.; Yao, X.-B.; Zou, B.-S.; Song, Z.-Y.; Yang, A.O.Y.; Luo, R.-X.; liu, Y.; et al. Treatment with Arbidol and Moxifloxacin in Ordinary and Severe Adult Patients Infected with COVID-19. medRxiv 2020. [Google Scholar] [CrossRef]
- Fan, L.; Liu, H.; Li, N.; Liu, C.; Gu, Y.; Liu, Y.; Chen, Y. Medical treatment of 55 patients with COVID-19 from seven cities in northeast China who fully recovered: A single-center, retrospective, observational study. Medicine 2021, 100, e23923. [Google Scholar] [CrossRef] [PubMed]
- Teoh, S.L.; Lim, Y.H.; Lai, N.M.; Lee, S.W. Directly acting antivirals for COVID-19: Where do we stand? Front. Microbiol. 2020, 11, 1857. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Yu, K.; Wang, Y.; Xu, W.; Ma, H.; Hou, Y.; Li, Y.; Cai, B.; Zhu, L.; Zhang, M. Efficacy and safety of triazavirin therapy for coronavirus disease 2019: A pilot randomized controlled trial. Engineering 2020, 6, 1185–1191. [Google Scholar] [CrossRef]
- Huang, D.; Yu, H.; Wang, T.; Yang, H.; Yao, R.; Liang, Z. Efficacy and safety of umifenovir for coronavirus disease 2019 (COVID-19): A systematic review and meta-analysis. J. Med. Virol. 2021, 93, 481–490. [Google Scholar] [CrossRef]
- Furuta, Y.; Komeno, T.; Nakamura, T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn. Acad. Ser. B 2017, 93, 449–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagata, T.; Lefor, A.K.; Hasegawa, M.; Ishii, M. Favipiravir: A new medication for the Ebola virus disease pandemic. Disaster Med. Public Health Prep. 2015, 9, 79–81. [Google Scholar] [CrossRef]
- Cai, Q.; Yang, M.; Liu, D.; Chen, J.; Shu, D.; Xia, J.; Liao, X.; Gu, Y.; Cai, Q.; Yang, Y. Experimental treatment with favipiravir for COVID-19: An open-label control study. Engineering 2020, 6, 1192–1198. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, A.; Mostafa, A.; Al-Karmalawy, A.A.; Zidan, A.; Abulkhair, H.S.; Mahmoud, S.H.; Shehata, M.; Elhefnawi, M.M.; Ali, M.A. Telaprevir is a potential drug for repurposing against SARS-CoV-2: Computational and in vitro studies. Heliyon 2021, 7, e07962. [Google Scholar] [CrossRef] [PubMed]
- Al-Karmalawy, A.A.; Alnajjar, R.; Dahab, M.; Metwaly, A.; Eissa, I. Molecular docking and dynamics simulations reveal the potential of anti-HCV drugs to inhibit COVID-19 main protease. Pharm Sci. 2021, 27, S109–S121. [Google Scholar] [CrossRef]
- Grein, J.; Ohmagari, N.; Shin, D.; Diaz, G.; Asperges, E.; Castagna, A.; Feldt, T.; Green, G.; Green, M.L.; Lescure, F.-X. Compassionate use of remdesivir for patients with severe Covid-19. N. Engl. J. Med. 2020, 382, 2327–2336. [Google Scholar] [CrossRef]
- Gordon, C.J.; Tchesnokov, E.P.; Schinazi, R.F.; Gotte, M. Molnupiravir promotes SARS-CoV-2 mutagenesis via the RNA template. J. Biol. Chem. 2021, 297, 100770. [Google Scholar] [CrossRef]
- Padhi, A.K.; Shukla, R.; Saudagar, P.; Tripathi, T. High-throughput rational design of the remdesivir binding site in the RdRp of SARS-CoV-2: Implications for potential resistance. iScience 2021, 24, 101992. [Google Scholar] [CrossRef] [PubMed]
- Robson, F.; Khan, K.S.; Le, T.K.; Paris, C.; Demirbag, S.; Barfuss, P.; Rocchi, P.; Ng, W.L. Coronavirus RNA Proofreading: Molecular Basis and Therapeutic Targeting. Mol. Cell 2020, 80, 1136–1138. [Google Scholar] [CrossRef]
- Pruijssers, A.J.; Denison, M.R. Nucleoside analogues for the treatment of coronavirus infections. Curr. Opin. Virol. 2019, 35, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Wahl, A.; Gralinski, L.E.; Johnson, C.E.; Yao, W.; Kovarova, M.; Dinnon, K.H.; Liu, H.; Madden, V.J.; Krzystek, H.M.; De, C.; et al. SARS-CoV-2 infection is effectively treated and prevented by EIDD-2801. Nature 2021, 591, 451–457. [Google Scholar] [CrossRef]
- Sheahan, T.P.; Sims, A.C.; Zhou, S.; Graham, R.L.; Pruijssers, A.J.; Agostini, M.L.; Leist, S.R.; Schäfer, A.; Dinnon, K.H., 3rd; Stevens, L.J.; et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020, 12, 541. [Google Scholar] [CrossRef] [Green Version]
- Abdelnabi, R.; Foo, C.S.; De Jonghe, S.; Maes, P.; Weynand, B.; Neyts, J. Molnupiravir Inhibits Replication of the Emerging SARS-CoV-2 Variants of Concern in a Hamster Infection Model. J. Infect. Dis. 2021, 224, 749–753. [Google Scholar] [CrossRef]
- Lee, C.-C.; Hsieh, C.-C.; Ko, W.-C. Molnupiravir—A Novel Oral Anti-SARS-CoV-2 Agent. Antibiotics 2021, 10, 1294. [Google Scholar] [CrossRef]
- Mahase, E. Covid-19: Molnupiravir reduces risk of hospital admission or death by 50% in patients at risk, MSD reports. BMJ 2021, 375, n2422. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Guo, L.; Chen, L.; Liu, W.; Cao, Y.; Zhang, J.; Feng, L. A case report of neonatal COVID-19 infection in China. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2020, 71, 853–857. [Google Scholar] [CrossRef]
- Cao, B.; Wang, Y.; Wen, D.; Liu, W.; Wang, J.; Fan, G.; Ruan, L.; Song, B.; Cai, Y.; Wei, M. A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020, 382, 1787–1799. [Google Scholar] [CrossRef]
- Del Amo, J.; Polo, R.; Moreno, S.; Díaz, A.; Martínez, E.; Arribas, J.R.; Jarrín, I.; Hernán, M.A. Incidence and severity of COVID-19 in HIV-positive persons receiving antiretroviral therapy: A cohort study. Ann. Intern. Med. 2020, 173, 536–541. [Google Scholar] [CrossRef]
- Tong, S.; Su, Y.; Yu, Y.; Wu, C.; Chen, J.; Wang, S.; Jiang, J. Ribavirin therapy for severe COVID-19: A retrospective cohort study. Int. J. Antimicrob. Agents 2020, 56, 106114. [Google Scholar] [CrossRef]
- Yang, G.; Tan, Z.; Zhou, L.; Yang, M.; Peng, L.; Liu, J.; Cai, J.; Yang, R.; Han, J.; Huang, Y. Effects of angiotensin II receptor blockers and ACE (angiotensin-converting enzyme) inhibitors on virus infection, inflammatory status, and clinical outcomes in patients with COVID-19 and hypertension: A single-center retrospective study. Hypertension 2020, 76, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Chen, Z.; Ni, L.; Chen, L.; Zhou, C.; Gao, C.; Wu, X.; Hua, L.; Huang, X.; Cui, X.; et al. Impact of Angiotensin-Converting Enzyme Inhibitors and Angiotensin Receptor Blockers on the Inflammatory Response and Viral Clearance in COVID-19 Patients. Front. Cardiovasc. Med. 2021, 8, 937. [Google Scholar] [CrossRef]
- Sanders, J.M.; Monogue, M.L.; Jodlowski, T.Z.; Cutrell, J.B. Pharmacologic Treatments for Coronavirus Disease 2019 (COVID-19): A Review. JAMA 2020, 323, 1824–1836. [Google Scholar] [CrossRef]
- Diaz-Arocutipa, C.; Saucedo-Chinchay, J.; Hernandez, A.V. Association Between ACEIs or ARBs Use and Clinical Outcomes in COVID-19 Patients: A Systematic Review and Meta-analysis. medRxiv 2020. [Google Scholar] [CrossRef]
- Li, M.; Wang, Y.; Ndiwane, N.; Orner, M.B.; Palacios, N.; Mittler, B.; Berlowitz, D.; Kazis, L.E.; Xia, W. The association of COVID-19 occurrence and severity with the use of angiotensin converting enzyme inhibitors or angiotensin-II receptor blockers in patients with hypertension. PLoS ONE 2021, 16, e0248652. [Google Scholar] [CrossRef] [PubMed]
- Soleimani, A.; Kazemian, S.; Karbalai Saleh, S.; Aminorroaya, A.; Shajari, Z.; Hadadi, A.; Talebpour, M.; Sadeghian, H.; Payandemehr, P.; Sotoodehnia, M. Effects of angiotensin receptor blockers (ARBs) on in-hospital outcomes of patients with hypertension and confirmed or clinically suspected COVID-19. Am. J. Hypertens. 2020, 33, 1102–1111. [Google Scholar] [CrossRef]
- European Society of Cardiology. First Randomised Trial Backs Safety of Common Heart Drugs in COVID-19 Patients: BRACE CORONA Trial Presented in a Hot Line Session today at ESC Congress 2020. ScienceDaily. 2020. Available online: https://www.sciencedaily.com/releases/2020/09/200901112216.htm. (accessed on 10 October 2021).
- Derington, C.G.; Cohen, J.B.; Mohanty, A.F.; Greene, T.H.; Cook, J.; Ying, J.; Wei, G.; Herrick, J.S.; Stevens, V.W.; Jones, B.E. Angiotensin II receptor blocker or angiotensin-converting enzyme inhibitor use and COVID-19-related outcomes among US Veterans. PLoS ONE 2021, 16, e0248080. [Google Scholar] [CrossRef]
- Hippisley-Cox, J.; Young, D.; Coupland, C.; Channon, K.M.; Tan, P.S.; Harrison, D.A.; Rowan, K.; Aveyard, P.; Pavord, I.D.; Watkinson, P.J. Risk of severe COVID-19 disease with ACE inhibitors and angiotensin receptor blockers: Cohort study including 8.3 million people. Heart 2020, 106, 1503. [Google Scholar] [CrossRef] [PubMed]
- Coenen, S.; van Der Velden, A.W.; Cianci, D.; Goossens, H.; Bongard, E.; Saville, B.R.; Gobat, N.; de Paor, M.; Ieven, M.; Verheij, T.J. Oseltamivir for coronavirus illness: Post-hoc exploratory analysis of an open-label, pragmatic, randomised controlled trial in European primary care from 2016 to 2018. Br. J. Gen. Pract. 2020, 70, e444–e449. [Google Scholar] [CrossRef]
- Lou, Y.; Liu, L.; Yao, H.; Hu, X.; Su, J.; Xu, K.; Luo, R.; Yang, X.; He, L.; Lu, X. Clinical outcomes and plasma concentrations of baloxavir marboxil and favipiravir in COVID-19 patients: An exploratory randomized, controlled trial. Eur. J. Pharm. Sci. 2021, 157, 105631. [Google Scholar] [CrossRef] [PubMed]
- Turtle, C.J.; Hay, K.A.; Gust, J.; Hanafi, L.-A.; Li, D.; Liles, W.C.; Wurfel, M.; Harju-Baker, S.; Myerson, D.; Gonzalez-Cuyar, L.; et al. Cytokine release syndrome (CRS) and neurotoxicity (NT) after CD19-specific chimeric antigen receptor- (CAR-) modified T cells. J. Clin. Oncol. 2017, 35, 3020. [Google Scholar] [CrossRef]
- Sarosiek, S.; Shah, R.; Munshi, N.C. Review of siltuximab in the treatment of multicentric Castleman’s disease. Ther. Adv. Hematol. 2016, 7, 360–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.-H.; Lu, C.-H.; Wong, S.H.; Lin, L.-T. Update on Antiviral Strategies Against COVID-19: Unmet Needs and Prospects. Front. Immunol. 2021, 11, 616595. [Google Scholar] [CrossRef]
- Salama, C.; Han, J.; Yau, L.; Reiss, W.G.; Kramer, B.; Neidhart, J.D.; Criner, G.J.; Kaplan-Lewis, E.; Baden, R.; Pandit, L.; et al. Tocilizumab in Patients Hospitalized with Covid-19 Pneumonia. N. Engl. J. Med. 2020, 384, 20–30. [Google Scholar] [CrossRef]
- Hartikainen, T.S.; Sörensen, N.A.; Haller, P.M.; Goßling, A.; Lehmacher, J.; Zeller, T.; Blankenberg, S.; Westermann, D.; Neumann, J.T. Clinical application of the 4th Universal Definition of Myocardial Infarction. Eur. Heart J. 2020, 41, 2209–2216. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Han, M.; Li, T.; Sun, W.; Wang, D.; Fu, B.; Zhou, Y.; Zheng, X.; Yang, Y.; Li, X. Effective treatment of severe COVID-19 patients with tocilizumab. Proc. Natl. Acad. Sci. USA 2020, 117, 10970–10975. [Google Scholar] [CrossRef]
- Luo, P.; Liu, Y.; Qiu, L.; Liu, X.; Liu, D.; Li, J. Tocilizumab treatment in COVID-19: A single center experience. J. Med. Virol. 2020, 92, 814–818. [Google Scholar] [CrossRef] [PubMed]
- Khiali, S.; Khani, E.; Entezari-Maleki, T. A comprehensive review of tocilizumab in covid-19 acute respiratory distress syndrome. J. Clin. Pharmacol. 2020, 60, 1131–1146. [Google Scholar] [CrossRef]
- Zhao, H.-M.; Xie, Y.-X.; Wang, C. Recommendations for respiratory rehabilitation in adults with coronavirus disease 2019. Chin. Med. J. 2020, 133, 1595. [Google Scholar] [CrossRef]
- Schiff, M.H.; Kremer, J.M.; Jahreis, A.; Vernon, E.; Isaacs, J.D.; van Vollenhoven, R.F. Integrated safety in tocilizumab clinical trials. Arthritis Res. Ther. 2011, 13, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Le, R.Q.; Li, L.; Yuan, W.; Shord, S.S.; Nie, L.; Habtemariam, B.A.; Przepiorka, D.; Farrell, A.T.; Pazdur, R. FDA approval summary: Tocilizumab for treatment of chimeric antigen receptor T cell-induced severe or life-threatening cytokine release syndrome. Oncologist 2018, 23, 943. [Google Scholar] [CrossRef] [Green Version]
- Sciascia, S.; Aprà, F.; Baffa, A.; Baldovino, S.; Boaro, D.; Boero, R.; Bonora, S.; Calcagno, A.; Cecchi, I.; Cinnirella, G. Pilot prospective open, single-arm multicentre study on off-label use of tocilizumab in severe patients with COVID-19. Clin. Exp. Rheumatol. 2020, 38, 529–532. [Google Scholar]
- Davoudi-Monfared, E.; Rahmani, H.; Khalili, H.; Hajiabdolbaghi, M.; Salehi, M.; Abbasian, L.; Kazemzadeh, H.; Yekaninejad, M.S. A randomized clinical trial of the efficacy and safety of interferon β-1a in treatment of severe COVID-19. Antimicrob. Agents Chemother. 2020, 64, e01061-20. [Google Scholar] [CrossRef]
- Kuriya, B.; Cohen, M.D.; Keystone, E. Baricitinib in rheumatoid arthritis: Evidence-to-date and clinical potential. Ther. Adv. Musculoskelet. Dis. 2017, 9, 37–44. [Google Scholar] [CrossRef]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Titanji, B.K.; Farley, M.M.; Mehta, A.; Connor-Schuler, R.; Moanna, A.; Cribbs, S.K.; O’Shea, J.; DeSilva, K.; Chan, B.; Edwards, A. Use of baricitinib in patients with moderate to severe coronavirus disease 2019. Clin. Infect. Dis. 2021, 72, 1247–1250. [Google Scholar] [CrossRef] [PubMed]
- Richardson, P.; Griffin, I.; Tucker, C.; Smith, D.; Oechsle, O.; Phelan, A.; Rawling, M.; Savory, E.; Stebbing, J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet 2020, 395, e30. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Chen, C.; Hu, F.; Wang, J.; Zhao, Q.; Gale, R.P.; Liang, Y. Impact of corticosteroid therapy on outcomes of persons with SARS-CoV-2, SARS-CoV, or MERS-CoV infection: A systematic review and meta-analysis. Leukemia 2020, 34, 1503–1511. [Google Scholar] [CrossRef]
- Stockman, L.J.; Bellamy, R.; Garner, P. SARS: Systematic review of treatment effects. PLoS Med. 2006, 3, e343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elmaaty, A.A.; Alnajjar, R.; Hamed, M.I.; Khattab, M.; Khalifa, M.M.; Al-Karmalawy, A.A. Revisiting activity of some glucocorticoids as a potential inhibitor of SARS-CoV-2 main protease: Theoretical study. RSC Adv. 2021, 11, 10027–10042. [Google Scholar] [CrossRef]
- Lansbury, L.; Rodrigo, C.; Leonardi-Bee, J.; Nguyen-Van-Tam, J.; Lim, W.S. Corticosteroids as adjunctive therapy in the treatment of influenza. Cochrane Database Syst. Rev. 2019, 2. [Google Scholar] [CrossRef]
- Delaney, J.W.; Pinto, R.; Long, J.; Lamontagne, F.; Adhikari, N.K.; Kumar, A.; Marshall, J.C.; Cook, D.J.; Jouvet, P.; Ferguson, N.D. The influence of corticosteroid treatment on the outcome of influenza A (H1N1pdm09)-related critical illness. Crit. Care 2016, 20, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Alshahrani, M.S.; Sindi, A.; Alshamsi, F.; Al-Omari, A.; El Tahan, M.; Alahmadi, B.; Zein, A.; Khatani, N.; Al-Hameed, F.; Alamri, S. Extracorporeal membrane oxygenation for severe Middle East respiratory syndrome coronavirus. Ann. Intensive Care 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Lamontagne, F.; Rochwerg, B.; Lytvyn, L.; Guyatt, G.H.; Møller, M.H.; Annane, D.; Kho, M.E.; Adhikari, N.K.; Machado, F.; Vandvik, P.O. Corticosteroid therapy for sepsis: A clinical practice guideline. BMJ 2018, 362, k3284. [Google Scholar] [CrossRef] [Green Version]
- Wirz, Y.; Meier, M.A.; Bouadma, L.; Luyt, C.E.; Wolff, M.; Chastre, J.; Tubach, F.; Schroeder, S.; Nobre, V.; Annane, D. Effect of procalcitonin-guided antibiotic treatment on clinical outcomes in intensive care unit patients with infection and sepsis patients: A patient-level meta-analysis of randomized trials. Crit. Care 2018, 22, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villar, J.; Ferrando, C.; Martínez, D.; Ambrós, A.; Muñoz, T.; Soler, J.A.; Aguilar, G.; Alba, F.; González-Higueras, E.; Conesa, L.A. Dexamethasone treatment for the acute respiratory distress syndrome: A multicentre, randomised controlled trial. Lancet Respir. Med. 2020, 8, 267–276. [Google Scholar] [CrossRef]
- Lee, M.K.; Choi, J.; Park, B.; Kim, B.; Lee, S.J.; Kim, S.H.; Yong, S.J.; Choi, E.H.; Lee, W.Y. High flow nasal cannulae oxygen therapy in acute-moderate hypercapnic respiratory failure. Clin. Respir. J. 2018, 12, 2046–2056. [Google Scholar] [CrossRef] [PubMed]
- Farthing, M.; Albonico, M.; Bisoffi, Z.; Bundy, D.; Buonfrate, D.; Chiodini, P.; Katelaris, P.; Kelly, P.; Savioli, L.; Mair, A.L. World Gastroenterology Organisation Global Guidelines: Management of Strongyloidiasis February 2018—Compact Version. J. Clin. Gastroenterol. 2020, 54, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Valentini, R.; Dupont, J.; Fernandez, J.; Solimano, J.; Palizas, F.; Riveros, D.; Saul, P.; Dupont, L.; Medina, J.; Falasco, V. Convalescent plasma as potential therapy for severe COVID-19 pneumonia. medRxiv 2020. [Google Scholar] [CrossRef]
- Mornese Pinna, S.; Lupia, T.; Scabini, S.; Vita, D.; De Benedetto, I.; Gaviraghi, A.; Colasanto, I.; Varese, A.; Cattel, F.; De Rosa, F.G.; et al. Monoclonal antibodies for the treatment of COVID-19 patients: An umbrella to overcome the storm? Int. Immunopharmacol. 2021, 101, 108200. [Google Scholar] [CrossRef]
- Taylor, P.C.; Adams, A.C.; Hufford, M.M.; de la Torre, I.; Winthrop, K.; Gottlieb, R.L. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021, 21, 382–393. [Google Scholar] [CrossRef] [PubMed]
- Razonable, R.R.; Pawlowski, C.; O’Horo, J.C.; Arndt, L.L.; Arndt, R.; Bierle, D.M.; Borgen, M.D.; Hanson, S.N.; Hedin, M.C.; Lenehan, P.; et al. Casirivimab–Imdevimab treatment is associated with reduced rates of hospitalization among high-risk patients with mild to moderate coronavirus disease-19. EClinicalMedicine 2021, 40, 101102. [Google Scholar] [CrossRef]
- Gupta, A.; Gonzalez-Rojas, Y.; Juarez, E.; Crespo Casal, M.; Moya, J.; Falci, D.R.; Sarkis, E.; Solis, J.; Zheng, H.; Scott, N.; et al. Early Treatment for Covid-19 with SARS-CoV-2 Neutralizing Antibody Sotrovimab. N. Engl. J. Med. 2021. [Google Scholar] [CrossRef]
- Duke, T.; Graham, S.; Cherian, M.; Ginsburg, A.; English, M.; Howie, S.; Peel, D.; Enarson, P.; Wilson, I.; Were, W. Oxygen is an essential medicine: A call for international action [Unresolved issues]. Int. J. Tuberc. Lung Dis. 2010, 14, 1362–1368. [Google Scholar] [PubMed]
- Rojas-Reyes, M.X.; Rugeles, C.G.; Charry-Anzola, L.P. Oxygen therapy for lower respiratory tract infections in children between 3 months and 15 years of age. Cochrane Database Syst. Rev. 2014, 12. [Google Scholar] [CrossRef] [PubMed]
- Kandeil, A.; Mostafa, A.; Kutkat, O.; Moatasim, Y.; Al-Karmalawy, A.A.; Rashad, A.A.; Kayed, A.E.; Kayed, A.E.; El-Shesheny, R.; Kayali, G.; et al. Bioactive Polyphenolic Compounds Showing Strong Antiviral Activities against Severe Acute Respiratory Syndrome Coronavirus 2. Pathogens 2021, 10, 758. [Google Scholar] [CrossRef]
- Tito, A.; Colantuono, A.; Pirone, L.; Pedone, E.; Intartaglia, D.; Giamundo, G.; Conte, I.; Vitaglione, P.; Apone, F. Pomegranate Peel Extract as an Inhibitor of SARS-CoV-2 Spike Binding to Human ACE2 Receptor (in vitro): A Promising Source of Novel Antiviral Drugs. Front. Chem. 2021, 9, 638187. [Google Scholar] [CrossRef]
- Yousefi, M.; Sadriirani, M.; PourMahmoudi, A.; Mahmoodi, S.; Samimi, B.; Hosseinikia, M.; Saeedinezhad, Z.; Panahande, S.B. Effects of pomegranate juice (Punica Granatum) on inflammatory biomarkers and complete blood count in patients with COVID-19: A structured summary of a study protocol for a randomized clinical trial. Trials 2021, 22, 246. [Google Scholar] [CrossRef]
- Aucoin, M.; Cooley, K.; Saunders, P.R.; Carè, J.; Anheyer, D.; Medina, D.N.; Cardozo, V.; Remy, D.; Hannan, N.; Garber, A. The effect of Echinacea spp. on the prevention or treatment of COVID-19 and other respiratory tract infections in humans: A rapid review. Adv. Integr. Med. 2020, 7, 203–217. [Google Scholar] [CrossRef]
- Al-Karmalawy, A.A.; Farid, M.M.; Mostafa, A.; Ragheb, A.Y.; Mahmoud, S.H.; Shehata, M.; Shama, N.M.A.; GabAllah, M.; Mostafa-Hedeab, G.; Marzouk, M.M. Naturally Available Flavonoid Aglycones as Potential Antiviral Drug Candidates against SARS-CoV-2. Molecules 2021, 26, 6559. [Google Scholar] [CrossRef]
- Soltane, R.; Chrouda, A.; Mostafa, A.; Al-Karmalawy, A.A.; Chouaïb, K.; Dhahri, A.; Pashameah, R.A.; Alasiri, A.; Kutkat, O.; Shehata, M.; et al. Strong Inhibitory Activity and Action Modes of Synthetic Maslinic Acid Derivative on Highly Pathogenic Coronaviruses: COVID-19 Drug Candidate. Pathogens 2021, 10, 623. [Google Scholar] [CrossRef] [PubMed]
- Zaki, A.A.; Al-Karmalawy, A.A.; El-Amier, Y.A.; Ashour, A. Molecular docking reveals the potential of Cleome amblyocarpa isolated compounds to inhibit COVID-19 virus main protease. New J. Chem. 2020, 44, 16752–16758. [Google Scholar] [CrossRef]
- Jan, J.-T.; Cheng, T.-J.R.; Juang, Y.-P.; Ma, H.-H.; Wu, Y.-T.; Yang, W.-B.; Cheng, C.-W.; Chen, X.; Chou, T.-H.; Shie, J.-J.; et al. Identification of existing pharmaceuticals and herbal medicines as inhibitors of SARS-CoV-2 infection. Proc. Natl. Acad. Sci. USA 2021, 118, e2021579118. [Google Scholar] [CrossRef]
- Thomas, S.; Patel, D.; Bittel, B.; Wolski, K.; Wang, Q.; Kumar, A.; Il’Giovine, Z.J.; Mehra, R.; McWilliams, C.; Nissen, S.E.; et al. Effect of high-dose zinc and ascorbic acid supplementation vs usual care on symptom length and reduction among ambulatory patients with sars-cov-2 infection: The covid a to z randomized clinical trial. JAMA Network Open 2021, 4, e210369. [Google Scholar] [CrossRef] [PubMed]
- Nimavat, N.; Singh, S.; Singh, P.; Singh, S.K.; Sinha, N. Vitamin D deficiency and COVID-19: A case-control study at a tertiary care hospital in India. Ann. Med. Surg. 2021, 68, 102661. [Google Scholar] [CrossRef]
- Lakkireddy, M.; Gadiga, S.G.; Malathi, R.D.; Karra, M.L.; Raju, I.S.S.V.P.M.; Ragini; Chinapaka, S.; Baba, K.S.S.S.; Kandakatla, M. Impact of daily high dose oral vitamin D therapy on the inflammatory markers in patients with COVID 19 disease. Sci. Rep. 2021, 11, 10641. [Google Scholar] [CrossRef]



| Structural Proteins | ||
|---|---|---|
| Protein | Function | Refs. |
| S (spike glycoprotein) It is a 150 KDa transmembrane protein found exposed to the outer surface of the virus | It helps in virus attachment and entry by binding with surface ACE2 receptors on the host cell. It is proteolytically cleaved into S1, “which determines host-virus infection, and cellular tropism”, and S2 unit “which controls virus fusion and entry”. | [68] |
| E (small envelope glycoprotein) Smallest protein in the virus | It helps in viral particle production and maturation. | [69] |
| M (membrane glycoprotein) Most abundant virus protein | It determines the envelope shape of the viral particle and is crucial for viral assembly. | [69] |
| N (nucleocapsid protein) localized in the endoplasmic reticulum–Golgi region and highly phosphorylated | It is crucial in virus replication and cellular responses to virus infection. It can make structural changes, resulting in enhancing virus infection. | [70] |
| Nonstructural proteins | ||
| Nsp 1 and 3 | Promoting cellular breakdown and limiting the translation of the host’s RNA, inhibiting type-I IFN signaling, and interfering with the innate immunity of infected cells. | [71,72] |
| Nsp 2 | Binding to prohibition protein. | |
| Nsp 3 and 5 | Elevating the expression of cytokine and the viral polyprotein cleavage. | |
| Nsp 4 and 6 | Contributing to DMVs formation as a transmembrane scaffold protein. | |
| Nsp 7/8 complex | Viral RNA polymerase subunits. | |
| Nsp 9 | RNA binding protein phosphatase. | |
| Nsp 10, 16, and 14 | Stimulation of ExoN and 2-O-MT activity. | |
| Nsp 12 | The main viral RNA-dependent RNA polymerase subunit. | |
| Nsp 13 | RNA helicase, 50 triphosphatases. | |
| Nsp 14 | Proofreading of the viral genome. | |
| Nsp 15 | Viral endoribonuclease and chymotrypsin-like protease. | |
| Nsp 16 | Averting MDA5 recognition and blocking cellular innate immunity. | |
| Viral Component | Function | Refs. |
|---|---|---|
| Double membrane vesicles | Prevents recognition of the viral dsRNA by PRRs, which is an intermediate product, leads to efficient virus-induced type I interferon. | [111] |
| Nsp 1 | Block type-I INF via host translational machinery inactivation and interfere with the phosphorylation of STAT1. Consequently, these activities lead to increased virus spreading and pathogenicity. | [112] |
| Nsp 3 | It encodes macrodomains and papain-like protease (PLpro) (cleavage of Nsps) proteins. Both Nsp 3/PLpro proteins contribute to the escaping of SARS-CoV-2 from induced immune response and block IFN responses. | [113] |
| ORF3b | The protein encoded on this gene segment can interfere with the INF signaling pathway via targetting IRF3 and/or MAVs | [65,91] |
| ORF6 | Binds to cellular karyopherin-a2 and anchoring karyopherin-b1 on internal membranes; the protein encoded on this gene segment suppresses the JAK–STAT signaling pathway, preventing nuclear translocation of the transcription factor STAT1. | [114] |
| ORF8 | Mediates immune evasion through downregulating the expression of surface major histocompatibility complex class Ι (MHC-Ι) | [115] |
| Nsp 13–15 & Nsp 6 | Suppress primary interferon production and interferon signaling | [116] |
| Nsp 14 and Nsp 16 | Help in imitating the capping machinery of the host, because the viral SARS-CoV genome composition has a 50 cap less than the host cell mRNA, thus making immune system cells recognize it and induce an immune response easily. Nsp 14 starts cap formation and thereafter follows the viral RNAs cap modification by Nsp16. Consequently, the viral RNA looks similar to host cell mRNA and evades possible PRR recognition. | [117] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Karmalawy, A.A.; Soltane, R.; Abo Elmaaty, A.; Tantawy, M.A.; Antar, S.A.; Yahya, G.; Chrouda, A.; Pashameah, R.A.; Mustafa, M.; Abu Mraheil, M.; et al. Coronavirus Disease (COVID-19) Control between Drug Repurposing and Vaccination: A Comprehensive Overview. Vaccines 2021, 9, 1317. https://doi.org/10.3390/vaccines9111317
Al-Karmalawy AA, Soltane R, Abo Elmaaty A, Tantawy MA, Antar SA, Yahya G, Chrouda A, Pashameah RA, Mustafa M, Abu Mraheil M, et al. Coronavirus Disease (COVID-19) Control between Drug Repurposing and Vaccination: A Comprehensive Overview. Vaccines. 2021; 9(11):1317. https://doi.org/10.3390/vaccines9111317
Chicago/Turabian StyleAl-Karmalawy, Ahmed A., Raya Soltane, Ayman Abo Elmaaty, Mohamed A. Tantawy, Samar A. Antar, Galal Yahya, Amani Chrouda, Rami Adel Pashameah, Muhamad Mustafa, Mobarak Abu Mraheil, and et al. 2021. "Coronavirus Disease (COVID-19) Control between Drug Repurposing and Vaccination: A Comprehensive Overview" Vaccines 9, no. 11: 1317. https://doi.org/10.3390/vaccines9111317
APA StyleAl-Karmalawy, A. A., Soltane, R., Abo Elmaaty, A., Tantawy, M. A., Antar, S. A., Yahya, G., Chrouda, A., Pashameah, R. A., Mustafa, M., Abu Mraheil, M., & Mostafa, A. (2021). Coronavirus Disease (COVID-19) Control between Drug Repurposing and Vaccination: A Comprehensive Overview. Vaccines, 9(11), 1317. https://doi.org/10.3390/vaccines9111317