Electroseparation of Slaughterhouse By-Product: Antimicrobial Peptide Enrichment by pH Modification
Abstract
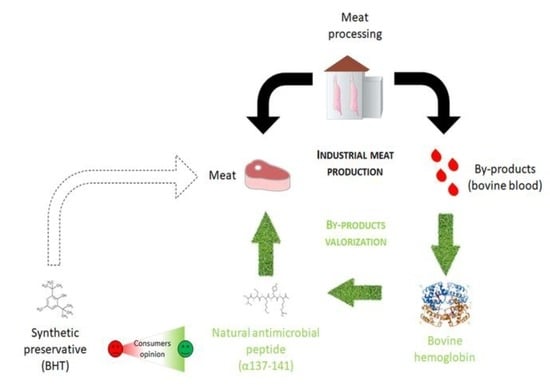
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Standard
2.2. Hydrolysis
2.3. Electroseparation
2.3.1. Equipment
2.3.2. Electroseparation Procedure
2.4. Analyses
2.4.1. RP-HPLC Analyses and Quantification Method for Total Peptides and α137–141 Peptide
2.4.2. Mass Spectrometry Analyses
3. Results and Discussion
3.1. Effect of pH Controlled at 4.7
3.1.1. Electrodialytic Parameter Evolution: pH, Conductivity, and Current Intensity
3.1.2. Total Peptide Migration Using pH Controlled at 4.7
3.1.3. α137-141 Recovery Using pH Control at 4.7
3.2. Improvement of the α137-141 Purity in the Recovery Compartment by pH Controlled at 6.5 and 9
3.2.1. Electrodialytic Parameter Evolution: Conductivity and Current Intensity
3.2.2. Total Peptide Migration by Controlling pH at 6.5 and 9
3.2.3. α137-141 Migration by Controlling pH at 6.5 and 9
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Castro, R.J.S.d.; Sato, H.H. Biologically active peptides: Processes for their generation, purification and identification and applications as natural additives in the food and pharmaceutical industries. Food Res. Int. 2015, 74, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, R.; Meisel, H. Food-derived peptides with biological activity: From research to food applications. Curr. Opin. Biotechnol. 2007, 18, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Li-Chan, E.C. Bioactive peptides and proteins hydrolysates: Research trends and challenges for application as nutraceuticals and functional food ingredients. Curr. Opin. Biotechnol. 2015, 1, 28–37. [Google Scholar] [CrossRef] [Green Version]
- Korhonen, H.; Pihlanto, A. Bioactive peptides: Production and functionality. Int. Dairy J. 2006, 16, 945–960. [Google Scholar] [CrossRef]
- Arroume, N.; Froidevaux, R.; Kapel, R.; Cudennec, B.; Ravallec, R.; Flahaut, C.; Bazinet, L.; Jacques, P.; Dhulster, P. Food peptides: Purification, identification and role in the metabolism. Curr. Opin. Food Sci. 2016, 7, 101–107. [Google Scholar] [CrossRef]
- Fernandez, A.; Riera, F.A. Influence of ionic strength on peptide membrane fractionation. Sep. Purif. Technol. 2013, 119, 129–135. [Google Scholar] [CrossRef]
- Kapel, R.; Froidevaux, R.; Nedjar-Arroume, N.; Fertin-Bazus, A.; Dhulster, P.; Guillochon, D. Continuous production of a peptidic fraction containing the intermediate opioid peptide LVV-haemorphin-7 (LVVh-7) by peptic hydrolysis of bovine haemoglobin in a continuous membrane reactor. Biotechnol. Appl. Biochem. 2003, 37, 317–324. [Google Scholar] [CrossRef]
- Agyei, D.; Danquah, M. Industrial-scale manufacturing of pharmaceutical-grade bioactive peptides. Biotechnol. Adv. 2011, 29, 272–277. [Google Scholar] [CrossRef]
- Saxena, A.; Tripathi, B.P.; Kumar, M.; Shadi, V.K. Membrane-based techniques for the separation and purification of proteins: An overview. Adv. Colloid Interface Sci. 2009, 145, 1–22. [Google Scholar] [CrossRef]
- Bazinet, L.; Amiot, J.; Poulin, J.F.; Tremblay, A.; Labbé, D. Process and System for Separation of Organic Charged Compounds. U.S. Patent US 10/591,238, 24 January 2008. [Google Scholar]
- Roblet, C.; Akhtar, M.J.; Mikhaylin, S.; Pilon, G.; Gill, T.; Marette, A.; Bazinet, L. Enhancement of glucose uptake in muscular cell by peptide fractions separated by electrodialysis with filtration membrane from salmon frame protein hydrolysate. J. Funct. Foods 2016, 22, 337–346. [Google Scholar] [CrossRef]
- Doyen, A.; Udenigwe, C.C.; Mitchell, P.; Marette, A.; Aluko, R.E.; Bazinet, L. Anti-diabetic and antihypertensive activities of two flaxseed protein hydrolysate fractions revealed following their simulatenous separation by electrodialysis with ultrafiltration membranes. Food Chem. 2014, 145, 66–76. [Google Scholar] [CrossRef]
- Bah, C.S.F.; Bekhit, A.E.-D.A.; Carne, A.; McConnell, M.A. Slaughterhouse blood: An emerging source of bioactive compounds. Compr. Rev. Food Sci. Food Saf. 2013, 12, 314–331. [Google Scholar] [CrossRef]
- Nedjar-Arroume, N.; Dubois-Delval, V.; Adje, E.Y.; Traisnel, J.; Krier, F.; Mary, P.; Kouach, M.; Briand, G.; Guillochon, D. Bovine hemoglobin: An attractive source of antibacterial peptides. Peptides 2008, 29, 969–977. [Google Scholar] [CrossRef] [PubMed]
- Catiau, L.; Traisnel, J.; Delval-Dubois, V.; Chihib, N.-E.; Guillochon, D.; Nedjar-Arroume, N. Minimal antimicrobial peptide sequence from hemoglobin alpha-chain: KYR. Peptides 2011, 32, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Przybylski, R.; Firdaous, L.; Châtaigné, G.; Dhulster, P.; Nedjar, N. Production of an antimicrobial peptide derived from slaughterhouse by-product and its potential application as meat preservative. Food Chem. 2016, 211, 306–313. [Google Scholar] [CrossRef]
- Przybylski, R.; Bazinet, L.; Firdaous, L.; Kouach, M.; Goossens, J.-F.; Dhulster, P.; Nedjar, N. Harnessing slaughterhouse by-products: From wastes to high-added value natural food preservative. Food Chem. 2020, 304, 125448. [Google Scholar] [CrossRef]
- Crosby, W.H.; Munn, J.L.; Furth, F.W. Standardizing a method for clinical hemoglobimetry. US Armed Forces Med. J. 1954, 5, 693–703. [Google Scholar]
- Dubois, V.; Nedjar-Arroume, N.; Guillochon, D. Influence of pH on the appearance of active peptides in the course of peptic hydrolysis of bovine haemoglobin. Prep. Biochem. Biotechnol. 2005, 35, 85–102. [Google Scholar] [CrossRef]
- Church, F.C.; Swaisgood, H.E.; Porter, D.H.; Catignani, L. Spectrophotometric assay using o-phthaldialdehyde for determination of proteolysis in milk and isolated milk proteins. J. Dairy Sci. 1983, 66, 1219–1227. [Google Scholar] [CrossRef]
- Vanhoute, M.; Firdaous, L.; Bazinet, L.; Froidevaux, R.; Lecouturier, D.; Guillochon, D.; Dhulster, P. Effect of haem on the fractionation of bovine haemoglobin peptic hydrolysate by electrodialysis with ultrafiltration membranes. J. Membr. Sci. 2010, 365, 16–24. [Google Scholar] [CrossRef]
- Doyen, A.; Roblet, C.; Beaulieu, L.; Saucier, L.; Pouliot, Y.; Bazinet, L. Impact of water splitting phenomenon during electrodialysis with ultrafiltration membranes on peptide selectivity and migration. J. Membr. Sci. 2013, 428, 349–356. [Google Scholar] [CrossRef]
- Poulin, J.F.; Amiot, J.; Bazinet, L. Improved peptide fractionation by electrodialysis with ultrafiltration membrane: Influence of ultrafiltration membrane stacking and electrical field strength. J. Membr. Sci. 2007, 299, 83–90. [Google Scholar] [CrossRef]
- Hedhili, K.; Vauchel, P.; Dimitrov, K.; Kriaa, K.; Châtaigné, G.; Hani, K.; Dhulster, P.; Nedjar-Arroume, N. Mechanism and kinetics modeling of the enzymatic hydrolysis of a1-32 antibacterial peptide. Bioprocess Biosyst. Eng. 2014, 37, 1315–1323. [Google Scholar] [CrossRef] [PubMed]
- Hedhili, K.; Dimitrov, K.; Vauchel, P.; Sila, A.; Chataigné, G.; Dhulster, P.; Nedjar, N. Valorization of cruor slaughterhouse by-product by enzymatic hydrolysis for the production of antibacterial peptides: Focus on α1-32 family peptides mechanism and kinectics modelling. Bioprocess Biosyst. Eng. 2015, 38, 1867–1877. [Google Scholar] [CrossRef] [PubMed]
- Adje, E.Y.; Balti, R.; Kouach, M.; Guillochon, D.; Nedjar-Arroume, N. α67-106 of bovine hemoglobin: A new family of antimicrobial and angiotensin I-converting enzyme inhibitory peptides. Eur. Food Res. Technol. 2011, 232, 637–646. [Google Scholar] [CrossRef]
- Froidevaux, R.; Nedjar-Arroume, N.; Choisnard, L.; Bigan, M.; Guillochon, D. Using an experimental design for the optimization of LVV-haemorphin-7 and VV-haemorphin-7 extraction by an organic solvent mixture in the course of bovine haemoglobin peptic hydrolysis. Biotechnol. Appl. Biochem. 2001, 33, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Lebrun, F.; Bazus, A.; Dhulster, P.; Guillochon, D. Influence of molecular interaction of bovine haemoglobin hydrolysate with an organic membrane. J. Membr. Sci. 1998, 146, 113–124. [Google Scholar] [CrossRef]
- Galier, S.; Balmann, H.R.-d. Electrophoretic membrane contactors. Chem. Eng. Res. Des. 2005, 83, 268–275. [Google Scholar] [CrossRef]
- Roblet, C.; Doyen, A.; Amiot, J.; Bazinet, L. Impact of pH on ultrafiltration membrane selectivity during EDUF purification of soy peptides from a complex matrix. J. Membr. Sci. 2013, 435, 207–217. [Google Scholar] [CrossRef]
- Gourley, L.; Gauthier, S.F.; Pouliot, Y.; Mollé, D.; Léonil, J.; Maubois, J.F. Identification of casein peptides interacting with polysulfone ultrafiltration membranes. Le Lait 1998, 78, 633–646. [Google Scholar] [CrossRef]
- Gourley, L.; Gauthier, S.F.; Pouliot, Y. Separation of casein hydrolysates using polysulfone ultrafiltration membranes with pH and EDTA treatments applied. Le Lait 1995, 75, 259–269. [Google Scholar] [CrossRef]
- Kadel, S.; Pellerin, G.; Thibodeau, J.; Perreault, V.; Lainé, C.; Bazinet, L. How molecular weight cut-offs and physicochemical properties of polyether sulfone membrane affect peptides migration and selectivity during electrodialysis with filtration membranes. Membranes 2019, 9, 153. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| pH | No Control | 4.7 | 6.5 | 9 |
|---|---|---|---|---|
| Total peptide concentration (mg·L−1) | 38.5 ± 5.5 | 108.4 ± 2.8 | 20.8 ± 12.5 | 8.2 ± 0.4 |
| Number of peptides | 16 | 40 | 23 | 6 |
| α137-141 concentration (mg·L−1) | 5.3 ± 0.3 | 10.5 ± 1.5 | 6.9 ± 2.4 | 4.9 ± 1.2 |
| α137-141 purity (%) | 10.3 ± 1.0 | 6.7 ± 0.9 | 37.2 ± 9.9 | 56.1 ± 11.3 |
| α137-141 enrichment factor | - | 9 | 50 | 75 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Przybylski, R.; Bazinet, L.; Firdaous, L.; Kouach, M.; Goossens, J.-F.; Dhulster, P.; Nedjar-Arroume, N. Electroseparation of Slaughterhouse By-Product: Antimicrobial Peptide Enrichment by pH Modification. Membranes 2020, 10, 90. https://doi.org/10.3390/membranes10050090
Przybylski R, Bazinet L, Firdaous L, Kouach M, Goossens J-F, Dhulster P, Nedjar-Arroume N. Electroseparation of Slaughterhouse By-Product: Antimicrobial Peptide Enrichment by pH Modification. Membranes. 2020; 10(5):90. https://doi.org/10.3390/membranes10050090
Chicago/Turabian StylePrzybylski, Rémi, Laurent Bazinet, Loubna Firdaous, Mostafa Kouach, Jean-François Goossens, Pascal Dhulster, and Naïma Nedjar-Arroume. 2020. "Electroseparation of Slaughterhouse By-Product: Antimicrobial Peptide Enrichment by pH Modification" Membranes 10, no. 5: 90. https://doi.org/10.3390/membranes10050090
APA StylePrzybylski, R., Bazinet, L., Firdaous, L., Kouach, M., Goossens, J. -F., Dhulster, P., & Nedjar-Arroume, N. (2020). Electroseparation of Slaughterhouse By-Product: Antimicrobial Peptide Enrichment by pH Modification. Membranes, 10(5), 90. https://doi.org/10.3390/membranes10050090