Evaluation of Cell-Free Synthesized Human Channel Proteins for In Vitro Channel Research
Abstract
:1. Introduction
2. Materials and Methods
2.1. Production of 8 Voltage-Gated Potassium Ion Channels by Bilayer Dialysis Method
2.2. Density Gradient Centrifugation
2.3. Single Channel Current Recording by PLB Assay
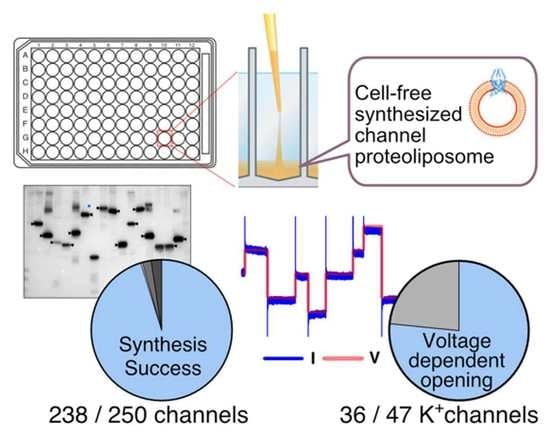
2.4. Cell-Free Synthesis of 250 Human Channels by Bilayer Method
2.5. In Vitro Co-Synthesis and Immunoprecipitation of KCNB1 and KCNS3
3. Results
3.1. Cell-Free Synthesis of KV and K2P Potassium Ion Channels
3.2. PLB Assay on Cell-Free Synthesized KCNK2
3.3. Cell-Free Synthesis of 250 Human Channel Proteins
3.4. Functional Evaluation of 47 Cell-Free Synthesized Voltage-Gated Potassium Ion Channels by PLB Assay
3.5. Cell-Free Synthesized KCNB1 and KCNS3 Form a Heterocomplex
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bagal, S.K.; Brown, A.D.; Cox, P.J.; Omoto, K.; Owen, R.M.; Pryde, D.C.; Sidders, B.; Skerratt, S.E.; Stevens, E.B.; Storer, R.I.; et al. Ion Channels as Therapeutic Targets: A Drug Discovery Perspective. J. Med. Chem. 2013, 56, 593–624. [Google Scholar] [CrossRef] [PubMed]
- Hutchings, C.J.; Colussi, P.; Clark, T.G. Ion Channels as Therapeutic Antibody Targets. MAbs 2019, 11, 265–296. [Google Scholar] [CrossRef] [PubMed]
- Kaczorowski, G.J.; McManus, O.B.; Priest, B.T.; Garcia, M.L. Ion Channels as Drug Targets: The next GPCRs. J. Gen. Physiol. 2008, 131, 399–405. [Google Scholar] [CrossRef] [PubMed]
- McGivern, J.G.; Ding, M. Ion Channels and Relevant Drug Screening Approaches. SLAS Discov 2020, 25, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.; Ursu, O.; Gaulton, A.; Bento, A.P.; Donadi, R.S.; Bologa, C.G.; Karlsson, A.; Al-Lazikani, B.; Hersey, A.; Oprea, T.I.; et al. A Comprehensive Map of Molecular Drug Targets. Nat. Rev. Drug Discov. 2017, 16, 19–34. [Google Scholar] [CrossRef]
- Guth, B.D. Preclinical Cardiovascular Risk Assessment in Modern Drug Development. Toxicol. Sci. 2007, 97, 4–20. [Google Scholar] [CrossRef] [Green Version]
- Nozawa, A.; Nanamiya, H.; Miyata, T.; Linka, N.; Endo, Y.; Weber, A.P.M.; Tozawa, Y. A Cell-Free Translation and Proteoliposome Reconstitution System for Functional Analysis of Plant Solute Transporters. Plant. Cell Physiol. 2007, 48, 1815–1820. [Google Scholar] [CrossRef] [Green Version]
- Nozawa, A.; Ogasawara, T.; Matsunaga, S.; Iwasaki, T.; Sawasaki, T.; Endo, Y. Production and Partial Purification of Membrane Proteins Using a Liposome-Supplemented Wheat Cell-Free Translation System. BMC Biotechnol. 2011, 11, 35. [Google Scholar] [CrossRef] [Green Version]
- Henrich, E.; Peetz, O.; Hein, C.; Laguerre, A.; Hoffmann, B.; Hoffmann, J.; Dotsch, V.; Bernhard, F.; Morgner, N. Analyzing Native Membrane Protein Assembly in Nanodiscs by Combined Non-Covalent Mass Spectrometry and Synthetic Biology. eLife Sci. 2017, 6, 243. [Google Scholar] [CrossRef]
- Dondapati, S.K.; Lübberding, H.; Zemella, A.; Thoring, L.; Wüstenhagen, D.A.; Kubick, S. Functional Reconstitution of Membrane Proteins Derived From Eukaryotic Cell-Free Systems. Front. Pharmacol. 2019, 10, 917. [Google Scholar] [CrossRef]
- Schoborg, J.A.; Hershewe, J.M.; Stark, J.C.; Kightlinger, W.; Kath, J.E.; Jaroentomeechai, T.; Natarajan, A.; DeLisa, M.P.; Jewett, M.C. A Cell-free Platform for Rapid Synthesis and Testing of Active Oligosaccharyltransferases. Biotechnol. Bioeng. 2018, 115, 739–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeda, H.; Ogasawara, T.; Ozawa, T.; Muraguchi, A.; Jih, P.-J.; Morishita, R.; Uchigashima, M.; Watanabe, M.; Fujimoto, T.; Iwasaki, T.; et al. Production of Monoclonal Antibodies against GPCR Using Cell-Free Synthesized GPCR Antigen and Biotinylated Liposome-Based Interaction Assay. Sci. Rep. 2015, 5, 11333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, W.; Takeda, H. Cell-Free Production of Proteoliposomes for Functional Analysis and Antibody Development Targeting Membrane Proteins. J. Vis. Exp. 2020, 163, e61871. [Google Scholar] [CrossRef] [PubMed]
- Jarecki, B.W.; Makino, S.; Beebe, E.T.; Fox, B.G.; Chanda, B. Function of Shaker Potassium Channels Produced by Cell-Free Translation upon Injection into Xenopus Oocytes. Sci. Rep. 2013, 3, 1040. [Google Scholar] [CrossRef] [Green Version]
- Friddin, M.S.; Smithers, N.P.; Beaugrand, M.; Marcotte, I.; Williamson, P.T.F.; Morgan, H.; Planque, M.R.R. de Single-Channel Electrophysiology of Cell-Free Expressed Ion Channels by Direct Incorporation in Lipid Bilayers. Analyst 2013, 138, 7294–7298. [Google Scholar] [CrossRef] [Green Version]
- Sackin, H.; Nanazashvili, M.; Makino, S. Direct Injection of Cell-Free Kir1.1 Protein into Xenopus Oocytes Replicates Single-Channel Currents Derived from Kir1.1 MRNA. Channels 2015, 9, 196–199. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Hasegawa, H.; Takemasa, E.; Suzuki, Y.; Oka, K.; Kiyoi, T.; Takeda, H.; Ogasawara, T.; Sawasaki, T.; Yasukawa, M.; et al. Efficiency and Safety of CRAC Inhibitors in Human Rheumatoid Arthritis Xenograft Models. J. Immunol. 2017, 199, 1584–1595. [Google Scholar] [CrossRef] [Green Version]
- Vaish, A.; Guo, S.; Murray, R.M.; Grandsard, P.J.; Chen, Q. On-Chip Membrane Protein Cell-Free Expression Enables Development of a Direct Binding Assay: A Curious Case of Potassium Channel KcsA-Kv1.3. Anal. Biochem. 2018, 556, 70–77. [Google Scholar] [CrossRef]
- Myshkin, M.Y.; Männikkö, R.; Krumkacheva, O.A.; Kulbatskii, D.S.; Chugunov, A.O.; Berkut, A.A.; Paramonov, A.S.; Shulepko, M.A.; Fedin, M.V.; Hanna, M.G.; et al. Cell-Free Expression of Sodium Channel Domains for Pharmacology Studies. Noncanonical Spider Toxin Binding Site in the Second Voltage-Sensing Domain of Human Nav1.4 Channel. Front. Pharmacol. 2019, 10, 953. [Google Scholar] [CrossRef]
- Montal, M.; Müeller, P. Formation of Bimolecular Membranes from Lipid Monolayers and a Study of Their Electrical Properties. Proc. Natl. Acad. Sci. USA 1972, 69, 3561–3566. [Google Scholar] [CrossRef]
- Zakharian, E. Recording of Ion Channel Activity in Planar Lipid Bilayer Experiments. Methods Mol. Biol. 2013, 998, 109–118. [Google Scholar] [PubMed] [Green Version]
- Strausberg, R.L.; Feingold, E.A.; Grouse, L.H.; Derge, J.G.; Klausner, R.D.; Collins, F.S.; Wagner, L.; Shenmen, C.M.; Schuler, G.D.; Altschul, S.F.; et al. Generation and Initial Analysis of More than 15,000 Full-Length Human and Mouse CDNA Sequences. Proc. Natl. Acad. Sci. USA 2002, 99, 16899–16903. [Google Scholar] [PubMed] [Green Version]
- Gibson, D.G. Enzymatic Assembly of Overlapping DNA Fragments. Meth. Enzymol. 2011, 498, 349–361. [Google Scholar]
- Nagase, T.; Yamakawa, H.; Tadokoro, S.; Nakajima, D.; Inoue, S.; Yamaguchi, K.; Itokawa, Y.; Kikuno, R.F.; Koga, H.; Ohara, O. Exploration of Human ORFeome: High-Throughput Preparation of ORF Clones and Efficient Characterization of Their Protein Products. DNA Res. 2008, 15, 137–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeda, H. Autoantibody Profiling Using Human Autoantigen Protein Array and AlphaScreen. Methods Mol. Biol. 2018, 1868, 93–112. [Google Scholar]
- Yano, T.; Takeda, H.; Uematsu, A.; Yamanaka, S.; Nomura, S.; Nemoto, K.; Iwasaki, T.; Takahashi, H.; Sawasaki, T. AGIA Tag System Based on a High Affinity Rabbit Monoclonal Antibody against Human Dopamine Receptor D1 for Protein Analysis. PLoS ONE 2016, 11, e0156716. [Google Scholar] [CrossRef] [Green Version]
- Bayburt, T.H.; Sligar, S.G. Membrane Protein Assembly into Nanodiscs. FEBS Lett. 2010, 584, 1721–1727. [Google Scholar] [CrossRef] [Green Version]
- Zúñiga, L.; Cayo, A.; González, W.; Vilos, C.; Zúñiga, R. Potassium Channels as a Target for Cancer Therapy: Current Perspectives. Oncotargets Ther. 2022, 15, 783–797. [Google Scholar] [CrossRef]
- Allen, N.M.; Weckhuysen, S.; Gorman, K.; King, M.D.; Lerche, H. Genetic Potassium Channel-Associated Epilepsies: Clinical Review of the Kv Family. Eur. J. Paediatr. Neurol. 2020, 24, 105–116. [Google Scholar] [CrossRef]
- González, C.; Baez-Nieto, D.; Valencia, I.; Oyarzún, I.; Rojas, P.; Naranjo, D.; Latorre, R. K(+) Channels: Function-Structural Overview. Compr. Physiol. 2012, 2, 2087–2149. [Google Scholar]
- Feliciangeli, S.; Chatelain, F.C.; Bichet, D.; Lesage, F. The Family of K2P Channels: Salient Structural and Functional Properties. J. Physiol. 2015, 593, 2587–2603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Connell, A.D.; Morton, M.J.; Hunter, M. Two-Pore Domain K+ Channels-Molecular Sensors. Biochim. Biophys. Acta 2002, 1566, 152–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maingret, F.; Lauritzen, I.; Patel, A.J.; Heurteaux, C.; Reyes, R.; Lesage, F.; Lazdunski, M.; Honoré, E. TREK-1 Is a Heat-activated Background K+ Channel. EMBO J. 2000, 19, 2483–2491. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.; Choe, C.; Kim, D. Thermosensitivity of the Two-pore Domain K+ Channels TREK-2 and TRAAK. J. Physiol. 2005, 564, 103–116. [Google Scholar] [CrossRef]
- Li, X.T.; Dyachenko, V.; Zuzarte, M.; Putzke, C.; Preisig-Müller, R.; Isenberg, G.; Daut, J. The Stretch-Activated Potassium Channel TREK-1 in Rat Cardiac Ventricular Muscle. Cardiovasc. Res. 2006, 69, 86–97. [Google Scholar]
- Alexander, S.P.H.; Mathie, A.; Peters, J.A.; Veale, E.L.; Striessnig, J.; Kelly, E.; Armstrong, J.F.; Faccenda, E.; Harding, S.D.; Pawson, A.J.; et al. THE CONCISE GUIDE TO PHARMACOLOGY 2021/22: Ion Channels. Br. J. Pharmacol. 2021, 178, S157–S245. [Google Scholar] [CrossRef]
- Tweedie, S.; Braschi, B.; Gray, K.; Jones, T.E.M.; Seal, R.L.; Yates, B.; Bruford, E.A. Genenames.Org: The HGNC and VGNC Resources in 2021. Nucleic Acids Res. 2020, 49, D939–D946. [Google Scholar] [CrossRef] [PubMed]
- Kerschensteiner, D.; Soto, F.; Stocker, M. Fluorescence Measurements Reveal Stoichiometry of K+ Channels Formed by Modulatory and Delayed Rectifier α-Subunits. Proc. Natl. Acad. Sci. USA 2005, 102, 6160–6165. [Google Scholar] [CrossRef] [Green Version]
- Patel, A.J.; Lazdunski, M.; Honoré, E. Kv2.1/Kv9.3, a Novel ATP-dependent Delayed-rectifier K+ Channel in Oxygen-sensitive Pulmonary Artery Myocytes. EMBO J. 1997, 16, 6615–6625. [Google Scholar] [CrossRef] [Green Version]
- Goshima, N.; Kawamura, Y.; Fukumoto, A.; Miura, A.; Honma, R.; Satoh, R.; Wakamatsu, A.; Yamamoto, J.; Kimura, K.; Nishikawa, T.; et al. Human Protein Factory for Converting the Transcriptome into an in Vitro–Expressed Proteome. Nat. Methods 2008, 5, 1011–1017. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Zhou, W.; Hamauchi, K.; Shirakura, K.; Doi, T.; Yagi, K.; Sawasaki, T.; Okada, Y.; Kondoh, M.; Takeda, H. Engineered Membrane Protein Antigens Successfully Induce Antibodies against Extracellular Regions of Claudin-5. Sci. Rep. 2018, 8, 8383. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.N.; Arenz, S.; Beckmann, R. Translation Regulation via Nascent Polypeptide-Mediated Ribosome Stalling. Curr. Opin. Struc. Biol. 2016, 37, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Khoubza, L.; Chatelain, F.C.; Feliciangeli, S.; Lesage, F.; Bichet, D. Physiological Roles of Heteromerization: Focus on the Two-pore Domain Potassium Channels. J. Physiol. 2021, 599, 1041–1055. [Google Scholar] [CrossRef] [PubMed]
- Bocksteins, E.; Snyders, D.J. Electrically Silent Kv Subunits: Their Molecular and Functional Characteristics. Physiology 2012, 27, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Goren, M.A.; Fox, B.G. Wheat Germ Cell-Free Translation, Purification, and Assembly of a Functional Human Stearoyl-CoA Desaturase Complex. Protein Expr. Purif. 2008, 62, 171–178. [Google Scholar] [CrossRef] [Green Version]
- Segawa, R.; Takeda, H.; Yokoyama, T.; Ishida, M.; Miyata, C.; Saito, T.; Ishihara, R.; Nakagita, T.; Sasano, Y.; Kanoh, N.; et al. A Chalcone Derivative Suppresses TSLP Induction in Mice and Human Keratinocytes through Binding to BET Family Proteins. Biochem. Pharm. 2021, 194, 114819. [Google Scholar] [CrossRef]
- Nishiyama, K.; Maekawa, M.; Nakagita, T.; Nakayama, J.; Kiyoi, T.; Chosei, M.; Murakami, A.; Kamei, Y.; Takeda, H.; Takada, Y.; et al. CNKSR1 Serves as a Scaffold to Activate an EGFR Phosphatase via Exclusive Interaction with RhoB-GTP. Life Sci. Alliance 2021, 4, e202101095. [Google Scholar] [CrossRef]
- Takahashi, H.; Yamanaka, S.; Kuwada, S.; Higaki, K.; Kido, K.; Sato, Y.; Fukai, S.; Tokunaga, F.; Sawasaki, T. A Human DUB Protein Array for Clarification of Linkage Specificity of Polyubiquitin Chain and Application to Evaluation of Its Inhibitors. Biomedicines 2020, 8, 152. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nishiguchi, R.; Tanaka, T.; Hayashida, J.; Nakagita, T.; Zhou, W.; Takeda, H. Evaluation of Cell-Free Synthesized Human Channel Proteins for In Vitro Channel Research. Membranes 2023, 13, 48. https://doi.org/10.3390/membranes13010048
Nishiguchi R, Tanaka T, Hayashida J, Nakagita T, Zhou W, Takeda H. Evaluation of Cell-Free Synthesized Human Channel Proteins for In Vitro Channel Research. Membranes. 2023; 13(1):48. https://doi.org/10.3390/membranes13010048
Chicago/Turabian StyleNishiguchi, Rei, Toyohisa Tanaka, Jun Hayashida, Tomoya Nakagita, Wei Zhou, and Hiroyuki Takeda. 2023. "Evaluation of Cell-Free Synthesized Human Channel Proteins for In Vitro Channel Research" Membranes 13, no. 1: 48. https://doi.org/10.3390/membranes13010048
APA StyleNishiguchi, R., Tanaka, T., Hayashida, J., Nakagita, T., Zhou, W., & Takeda, H. (2023). Evaluation of Cell-Free Synthesized Human Channel Proteins for In Vitro Channel Research. Membranes, 13(1), 48. https://doi.org/10.3390/membranes13010048