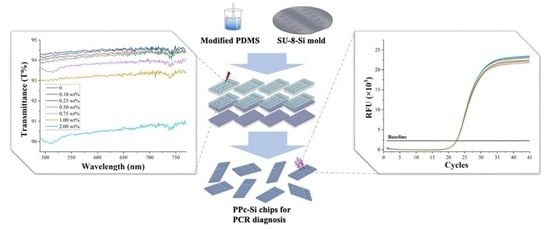
A Silicon-Based PDMS-PEG Copolymer Microfluidic Chip for Real-Time Polymerase Chain Reaction Diagnosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Fabrication of PPc-Si Chips
2.3. Optical Transmittance of the Patterned Slides for PPc-Si Chips
2.4. Contact Angle Monitoring of the Patterned Slides for PPc-Si Chips
2.5. Surface Morphology of the Reaction Channel’s Inner Surface
2.6. The Chemical State of the Reaction Channel’s Inner Surface
2.7. The Bonding Strength of the PPc-Si Chips
2.8. Real-Time PCR Test in PPc-Si Chips
3. Results and Discussion
3.1. Pattern Design and Fabrication of PPc-Si Chips
3.2. Optical Transmittance of PPSi Chips’ Reaction Channels
3.3. Surface Hydrophilicity of the Modified Patterned Slides
3.4. Surface Morphology of the Reaction Channel Inner Surface
3.5. Chemical State of the Reaction Channel Inner Surface
3.6. Bonding Strength of the PPc-Si Chips
3.7. Real-Time PCR Test in PPc-Si Chips
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Saiki, R.K.; Bugawan, T.L.; Horn, G.T.; Mullis, K.B.; Erlich, H.A. Analysis of enzymatically amplified β-globin and HLA-DQα DNA with allele-specific oligonucleotide probes. Nature 1986, 324, 163–166. [Google Scholar] [CrossRef]
- Saiki, R.; Gelfand, D.; Stoffel, S.; Scharf, S.; Higuchi, R.; Horn, G.; Mullis, K.; Erlich, H. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 1988, 239, 487–491. [Google Scholar] [CrossRef] [Green Version]
- Nicolini, U.; Lalatta, F.; Natacci, F.; Curcio, C.; Bui, T.-H. The introduction of QF-PCR in prenatal diagnosis of fetal aneuploidies: Time for reconsideration. Hum. Reprod. Update 2004, 10, 541–548. [Google Scholar] [CrossRef] [Green Version]
- Zimmermann, B.G.; Grill, S.; Holzgreve, W.; Zhong, X.Y.; Jackson, L.G.; Hahn, S. Digital PCR: A powerful new tool for noninvasive prenatal diagnosis? Prenat. Diagn. 2008, 28, 1087–1093. [Google Scholar] [CrossRef] [PubMed]
- Cirigliano, V.; Voglino, G.; Ordoñez, E.; Marongiu, A.; Paz Cañadas, M.; Ejarque, M.; Rueda, L.; Lloveras, E.; Fuster, C.; Adinolfi, M. Rapid prenatal diagnosis of common chromosome aneuploidies by QF-PCR, results of 9 years of clinical experience. Prenat. Diagn. 2009, 29, 40–49. [Google Scholar] [CrossRef]
- Saiki, R.; Scharf, S.; Faloona, F.; Mullis, K.; Horn, G.; Erlich, H.; Arnheim, N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 1985, 230, 1350–1354. [Google Scholar] [CrossRef]
- Jobling, M.A.; Gill, P. Encoded evidence: DNA in forensic analysis. Nat. Rev. Genet. 2004, 5, 739–751. [Google Scholar] [CrossRef] [PubMed]
- Welsh, J.; Petersen, C.; McClelland, M. Polymorphisms generated by arbitrarily primed PCR in the mouse: Application to strain identification and genetic mapping. Nucleic Acids Res. 1991, 19, 303–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Wang, L.; Mamon, H.; Kulke, M.H.; Berbeco, R.; Makrigiorgos, G.M. Replacing PCR with COLD-PCR enriches variant DNA sequences and redefines the sensitivity of genetic testing. Nat. Med. 2008, 14, 579–584. [Google Scholar] [CrossRef]
- Schaefer, B.C. Revolutions in rapid amplification of cDNA ends: New strategies for polymerase chain reaction cloning of full-length cDNA ends. Anal. Biochem. 1995, 227, 255–273. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, R.; Fockler, C.; Dollinger, G.; Watson, R. Kinetic PCR Analysis: Real-time Monitoring of DNA Amplification Reactions. Nat. Biotechnol. 1993, 11, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- Valasek, M.A.; Repa, J.J. The power of real-time PCR. Adv. Physiol. Educ. 2005, 29, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Heid, C.A.; Stevens, J.; Livak, K.J.; Williams, P.M. Real time quantitative PCR. Genome Res. 1996, 6, 986–994. [Google Scholar] [CrossRef] [Green Version]
- Cherpillod, P.; Schibler, M.; Vieille, G.; Cordey, S.; Mamin, A.; Vetter, P.; Kaiser, L. Ebola virus disease diagnosis by real-time RT-PCR: A comparative study of 11 different procedures. J. Clin. Virol. 2016, 77, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Xiao, A.T.; Tong, Y.X.; Zhang, S. Profile of RT-PCR for SARS-CoV-2: A Preliminary Study From 56 COVID-19 Patients. Clin. Infect. Dis. 2020, 71, 2249–2251. [Google Scholar] [CrossRef]
- Huggett, J.F.; French, D.; O’Sullivan, D.M.; Moran-Gilad, J.; Zumla, A. Monkeypox: Another test for PCR. Eurosurveillance 2022, 27, 2200497. [Google Scholar] [CrossRef]
- Gubala, V.; Harris, L.F.; Ricco, A.J.; Tan, M.X.; Williams, D.E. Point of Care Diagnostics: Status and Future. Anal. Chem. 2012, 84, 487–515. [Google Scholar] [CrossRef]
- Yang, S.; Wen, W. Lyophilized Ready-to-Use Mix for the Real-Time Polymerase Chain Reaction Diagnosis. ACS Appl. Bio Mater. 2021, 4, 4354–4360. [Google Scholar] [CrossRef]
- Manz, A.; Graber, N.; Widmer, H.M. Miniaturized total chemical analysis systems: A novel concept for chemical sensing. Sens. Actuators B Chem. 1990, 1, 244–248. [Google Scholar] [CrossRef]
- Reyes, D.R.; Iossifidis, D.; Auroux, P.-A.; Manz, A. Micro Total Analysis Systems. 1. Introduction, Theory, and Technology. Anal. Chem. 2002, 74, 2623–2636. [Google Scholar] [CrossRef]
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Wilding, P.; Shoffner, M.A.; Kricka, L.J. PCR in a silicon microstructure. Clin. Chem. 1994, 40, 1815–1818. [Google Scholar] [CrossRef]
- Kopp, M.U.; Mello, A.J.; Manz, A. Chemical amplification: Continuous-flow PCR on a chip. Science 1998, 280, 1046–1048. [Google Scholar] [CrossRef] [Green Version]
- Gerlach, A.; Maas, D.; Seidel, D.; Bartuch, H.; Schundau, S.; Kaschlik, K. Low-temperature anodic bonding of silicon to silicon wafers by means of intermediate glass layers. Microsyst. Technol. 1999, 5, 144–149. [Google Scholar] [CrossRef]
- Shi, X.; Lin, L.-I.; Chen, S.-Y.; Chao, S.-H.; Zhang, W.; Meldrum, D.R. Real-time PCR of single bacterial cells on an array of adhering droplets. Lab Chip 2011, 11, 2276. [Google Scholar] [CrossRef]
- Park, S.; Zhang, Y.; Lin, S.; Wang, T.-H.; Yang, S. Advances in microfluidic PCR for point-of-care infectious disease diagnostics. Biotechnol. Adv. 2011, 29, 830–839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Xing, D. Miniaturized PCR chips for nucleic acid amplification and analysis: Latest advances and future trends. Nucleic Acids Res. 2007, 35, 4223–4237. [Google Scholar] [CrossRef] [PubMed]
- Miranda, I.; Souza, A.; Sousa, P.; Ribeiro, J.; Castanheira, E.M.S.; Lima, R.; Minas, G. Properties and Applications of PDMS for Biomedical Engineering: A Review. J. Funct. Biomater. 2021, 13, 2. [Google Scholar] [CrossRef]
- Elenitoba-Johnson, O.; David, D.; Crews, N.; Wittwer, C.T. Plastic versus glass capillaries for rapid-cycle PCR. BioTechniques 2008, 44, 487–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Poll, M.L.; Zhou, F.; Ramstedt, M.; Hu, L.; Huck, W.T.S. A Self-Assembly Approach to Chemical Micropatterning of Poly(dimethylsiloxane). Angew. Chem. Int. Ed. 2007, 46, 6634–6637. [Google Scholar] [CrossRef]
- Barata, D.; Provaggi, E.; Van Blitterswijk, C.; Habibovic, P. Development of a microfluidic platform integrating high-resolution microstructured biomaterials to study cell–material interactions. Lab Chip 2017, 17, 4134–4147. [Google Scholar] [CrossRef]
- Morbioli, G.G.; Speller, N.C.; Stockton, A.M. A practical guide to rapid-prototyping of PDMS-based microfluidic devices: A tutorial. Anal. Chim. Acta 2020, 1135, 150–174. [Google Scholar] [CrossRef] [PubMed]
- Ongaro, A.E.; Di Giuseppe, D.; Kermanizadeh, A.; Miguelez Crespo, A.; Mencattini, A.; Ghibelli, L.; Mancini, V.; Wlodarczyk, K.L.; Hand, D.P.; Martinelli, E.; et al. Polylactic is a Sustainable, Low Absorption, Low Autofluorescence Alternative to Other Plastics for Microfluidic and Organ-on-Chip Applications. Anal. Chem. 2020, 92, 6693–6701. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.; Lee, L.P. Effects on wettability by surfactant accumulation/depletion in bulk polydimethylsiloxane (PDMS). Sens. Actuators B Chem. 2006, 119, 192–198. [Google Scholar] [CrossRef]
- Mukhopadhyay, R. When PDMS isn’t the best. Anal. Chem. 2007, 79, 3248–3253. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Fujii, T.; Nojima, T. PDMS–glass hybrid microreactor array with embedded temperature control device. Application to cell-free protein synthesis. Lab Chip 2002, 2, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Ranjit Prakash, A.; Adamia, S.; Sieben, V.; Pilarski, P.; Pilarski, L.M.; Backhouse, C.J. Small volume PCR in PDMS biochips with integrated fluid control and vapour barrier. Sens. Actuators B Chem. 2006, 113, 398–409. [Google Scholar] [CrossRef]
- Wu, W.; Loan Trinh, K.T.; Lee, N.Y. Flow-through PCR on a 3D qiandu-shaped polydimethylsiloxane (PDMS) microdevice employing a single heater: Toward microscale multiplex PCR. Analyst 2012, 137, 2069–2076. [Google Scholar] [CrossRef]
- Young Shik, S.; Keunchang, C.; Sun Hee, L.; Seok, C.; Sung-Jin, P.; Chanil, C.; Dong-Chul, H.; Jun Keun, C. PDMS-based micro PCR chip with Parylene coating. J. Micromech. Microeng. 2003, 13, 768. [Google Scholar] [CrossRef]
- Sivakumarasamy, R.; Nishiguchi, K.; Fujiwara, A.; Vuillaume, D.; Clément, N. A simple and inexpensive technique for PDMS/silicon chip alignment with sub-μm precision. Anal. Methods 2014, 6, 97–101. [Google Scholar] [CrossRef]
- Wolf, M.; Juncker, D.; Michel, B.; Hunziker, P.; Delamarche, E. Simultaneous detection of C-reactive protein and other cardiac markers in human plasma using micromosaic immunoassays and self-regulating microfluidic networks. Biosens. Bioelectron. 2004, 19, 1193–1202. [Google Scholar] [CrossRef]
- Li, S.; Floriano, P.N.; Christodoulides, N.; Fozdar, D.Y.; Shao, D.; Ali, M.F.; Dharshan, P.; Mohanty, S.; Neikirk, D.; McDevitt, J.T.; et al. Disposable polydimethylsiloxane/silicon hybrid chips for protein detection. Biosens. Bioelectron. 2005, 21, 574–580. [Google Scholar] [CrossRef]
- Toepke, M.W.; Beebe, D.J. PDMS absorption of small molecules and consequences in microfluidic applications. Lab Chip 2006, 6, 1484. [Google Scholar] [CrossRef] [PubMed]
- Dabaghi, M.; Shahriari, S.; Saraei, N.; Da, K.; Chandiramohan, A.; Selvaganapathy, P.R.; Hirota, J.A. Surface Modification of PDMS-Based Microfluidic Devices with Collagen Using Polydopamine as a Spacer to Enhance Primary Human Bronchial Epithelial Cell Adhesion. Micromachines 2021, 12, 132. [Google Scholar] [CrossRef]
- Cosson, S.; Lutolf, M. Hydrogel microfluidics for the patterning of pluripotent stem cells. Sci. Rep. 2014, 4, 4462. [Google Scholar] [CrossRef] [Green Version]
- Clancy, A.; Chen, D.; Bruns, J.; Nadella, J.; Stealey, S.; Zhang, Y.; Timperman, A.; Zustiak, S.P. Hydrogel-based microfluidic device with multiplexed 3D in vitro cell culture. Sci. Rep. 2022, 12, 17781. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Ellis, A.V.; Voelcker, N.H. Recent developments in PDMS surface modification for microfluidic devices. Electrophoresis 2010, 31, 2–16. [Google Scholar] [CrossRef] [PubMed]
- Hemmilä, S.; Cauich-Rodríguez, J.V.; Kreutzer, J.; Kallio, P. Rapid, simple, and cost-effective treatments to achieve long-term hydrophilic PDMS surfaces. Appl. Surf. Sci. 2012, 258, 9864–9875. [Google Scholar] [CrossRef]
- Sivakumar, R.; Lee, N.Y. Microfluidic device fabrication mediated by surface chemical bonding. Analyst 2020, 145, 4096–4110. [Google Scholar] [CrossRef] [PubMed]
- Nistorescu, S.; Icriverzi, M.; Florian, P.; Bonciu, A.; Marascu, V.; Dumitrescu, N.; Pircalabioru, G.G.; Rusen, L.; Mocanu, A.; Roseanu, A.; et al. Mitigation of Cellular and Bacterial Adhesion on Laser Modified Poly (2-Methacryloyloxyethyl Phosphorylcholine)/Polydimethylsiloxane Surface. Nanomaterials 2022, 13, 64. [Google Scholar] [CrossRef]
- Eddings, M.A.; Johnson, M.A.; Gale, B.K. Determining the optimal PDMS–PDMS bonding technique for microfluidic devices. J. Micromech. Microeng. 2008, 18, 067001. [Google Scholar] [CrossRef]
- Xiao, Y.; Yu, X.D.; Xu, J.J.; Chen, H.Y. Bulk modification of PDMS microchips by an amphiphilic copolymer. Electrophoresis 2007, 28, 3302–3307. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yan, H.; Ren, K.; Dai, W.; Wu, H. Convenient Method for Modifying Poly(dimethylsiloxane) with Poly(ethylene glycol) in Microfluidics. Anal. Chem. 2009, 81, 6627–6632. [Google Scholar] [CrossRef]
- Yao, M.; Fang, J. Hydrophilic PEO-PDMS for microfluidic applications. J. Micromech. Microeng. 2012, 22, 025012. [Google Scholar] [CrossRef]
- Hisyam, A.; Razak, A.; Szabo, P.; Skov, A.L. Enhancement of dielectric permittivity by incorporating PDMS-PEG multiblock copolymers in silicone elastomers. RSC Adv. 2015, 5, 53054–53062. [Google Scholar] [CrossRef] [Green Version]
- Scofield, J.M.P.; Gurr, P.A.; Kim, J.; Fu, Q.; Halim, A.; Kentish, S.E.; Qiao, G.G. High-performance thin film composite membranes with well-defined poly(dimethylsiloxane)-b-poly(ethylene glycol) copolymer additives for CO2 separation. J. Polym. Sci. Part A Polym. Chem. 2015, 53, 1500–1511. [Google Scholar] [CrossRef]
- Gökaltun, A.; Kang, Y.B.; Yarmush, M.L.; Usta, O.B.; Asatekin, A. Simple Surface Modification of Poly(dimethylsiloxane) via Surface Segregating Smart Polymers for Biomicrofluidics. Sci. Rep. 2019, 9, 7377. [Google Scholar] [CrossRef] [Green Version]
- Mair, D.B.; Williams, M.A.C.; Chen, J.F.; Goldstein, A.; Wu, A.; Lee, P.H.U.; Sniadecki, N.J.; Kim, D.-H. PDMS–PEG Block Copolymer and Pretreatment for Arresting Drug Absorption in Microphysiological Devices. ACS Appl. Mater. Interfaces 2022, 14, 38541–38549. [Google Scholar] [CrossRef] [PubMed]
- Borók, A.; Laboda, K.; Bonyár, A. PDMS Bonding Technologies for Microfluidic Applications: A Review. Biosensors 2021, 11, 292. [Google Scholar] [CrossRef]
- Yang, S.; Zhang, Z.; Xian, Q.; Song, Q.; Liu, Y.; Gao, Y.; Wen, W. An Aluminum-Based Microfluidic Chip for Polymerase Chain Reaction Diagnosis. Molecules 2023, 28, 1085. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Kodzius, R.; Xiao, K.; Qin, J.; Wen, W. Fast detection of genetic information by an optimized PCR in an interchangeable chip. Biomed. Microdevices 2012, 14, 179–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Q.; Sun, X.; Dai, Z.; Gao, Y.; Gong, X.; Zhou, B.; Wu, J.; Wen, W. Point-of-care testing detection methods for COVID-19. Lab Chip 2021, 21, 1634–1660. [Google Scholar] [CrossRef]
- Tichopad, A.; Dilger, M.; Schwarz, G.; Pfaffl, M.W. Standardized determination of real-time PCR efficiency from a single reaction set-up. Nucleic Acids Res. 2003, 31, e122. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Dorak, M.T. Real-Time PCR: Practical Issues and Troubleshooting; MOBGAM: Istanbul, Turkey, 2011. [Google Scholar]
- Kralik, P.; Ricchi, M. A Basic Guide to Real Time PCR in Microbial Diagnostics: Definitions, Parameters, and Everything. Front. Microbiol. 2017, 8, 108. [Google Scholar] [CrossRef] [PubMed] [Green Version]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, S.; Xian, Q.; Liu, Y.; Zhang, Z.; Song, Q.; Gao, Y.; Wen, W. A Silicon-Based PDMS-PEG Copolymer Microfluidic Chip for Real-Time Polymerase Chain Reaction Diagnosis. J. Funct. Biomater. 2023, 14, 208. https://doi.org/10.3390/jfb14040208
Yang S, Xian Q, Liu Y, Zhang Z, Song Q, Gao Y, Wen W. A Silicon-Based PDMS-PEG Copolymer Microfluidic Chip for Real-Time Polymerase Chain Reaction Diagnosis. Journal of Functional Biomaterials. 2023; 14(4):208. https://doi.org/10.3390/jfb14040208
Chicago/Turabian StyleYang, Siyu, Qingyue Xian, Yiteng Liu, Ziyi Zhang, Qi Song, Yibo Gao, and Weijia Wen. 2023. "A Silicon-Based PDMS-PEG Copolymer Microfluidic Chip for Real-Time Polymerase Chain Reaction Diagnosis" Journal of Functional Biomaterials 14, no. 4: 208. https://doi.org/10.3390/jfb14040208
APA StyleYang, S., Xian, Q., Liu, Y., Zhang, Z., Song, Q., Gao, Y., & Wen, W. (2023). A Silicon-Based PDMS-PEG Copolymer Microfluidic Chip for Real-Time Polymerase Chain Reaction Diagnosis. Journal of Functional Biomaterials, 14(4), 208. https://doi.org/10.3390/jfb14040208