Extracellular Synthesis and Characterization of Silver Nanoparticles—Antibacterial Activity against Multidrug-Resistant Bacterial Strains
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Materials
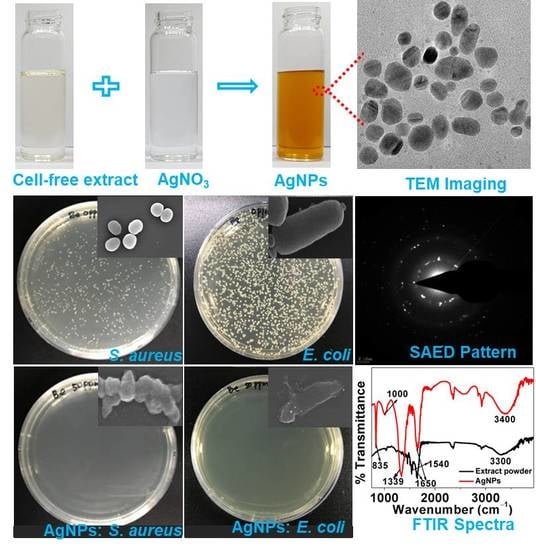
2.2. Extracellular Synthesis of AgNPs
2.3. Characterization of AgNPs
2.4. Antimicrobial Susceptibility Assay: Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC)
2.5. Optical Microscopy
2.6. Field-Emission Scanning Electron Microscopy
3. Results and Discussion
3.1. Effect of Cell-Free Beef Extract Concentration
3.2. Effect of NaOH Concentration
3.3. Effect of AgNO3 Concentration
3.4. Characterization of AgNPs
3.5. Stability Studies
3.6. Antibacterial Results
3.7. Bactericidal Concentration
3.8. Optical Microscopy
3.9. Field-Emission Scanning Electron Microscopy
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Foster, T.J. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol. Rev. 2017, 41, 430–449. [Google Scholar] [CrossRef]
- Mainil, J. Escherichia coli virulence factors. Vet. Immunol. Immunop. 2013, 152, 2–12. [Google Scholar] [CrossRef] [Green Version]
- Angulo-Pineda, C.; Palma, P.; Bejarano, J.; Riveros, A.; Kogan, M.; Palza, H. Antibacterial silver nanoparticles supported on graphene oxide with reduced cytotoxicity. JOM Miner. Met. Mater. Soc. 2019, 71, 3698–3705. [Google Scholar] [CrossRef]
- Duval, R.E.; Grare, M.; Demoré, B. Fight against antimicrobial resistance: We always need new antibacterials but for right bacteria. Molecules 2019, 24, 3152. [Google Scholar] [CrossRef] [Green Version]
- Subbiah, M.; Caudell, M.A.; Mair, C.; Davis, M.A.; Matthews, L.; Quinlan, R.J.; Quinlan, M.B.; Lyimo, B.; Buza, J.; Keyyu, J.; et al. Antimicrobial resistant enteric bacteria are widely distributed amongst people, animals and the environment in Tanzania. Nat. Commun. 2020, 11, 228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Remzova, M.; Zouzelka, R.; Brzicova, T.; Vrbova, K.; Pinkas, D.; Rőssner, P.; Topinka, J.; Rathousky, J. Toxicity of TiO2, ZnO and SiO2 nanoparticles in human lung cells: Safe-by-design development of construction materials. Nanomaterials 2019, 9, 968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seabra, A.B.; Durán, N. Nanotoxicology of metal oxide nanoparticles. Metals 2015, 5, 934–975. [Google Scholar] [CrossRef]
- Godymchuk, A.; Papina, I.; Karepina, E.; Kuznetsov, D.; Lapin, I.; Svetlichnyi, V. Agglomeration of iron oxide nanoparticles: pH effect is stronger than amino acid acidity. J. Nanopart. Res. 2019, 21, 208. [Google Scholar] [CrossRef]
- Zhao, Y.; Tian, Y.; Cui, Y.; Liu, W.; Ma, W.; Jiang, X. Small molecule-capped gold nanoparticles as potent antibacterial agents that target gram-negative bacteria. J. Am. Chem. Soc. 2010, 132, 12349–12356. [Google Scholar] [CrossRef]
- Li, X.; Robinson, S.M.; Gupta, A.; Saha, K.; Jiang, Z.; Moyano, D.F.; Sahar, A.; Riley, M.A.; Rotello, V.M. Functional gold nanoparticles as potent antimicrobial agents against multi-drug-resistant bacteria. ACS Nano 2014, 8, 10682–10686. [Google Scholar] [CrossRef]
- Pelgrift, R.Y.; Friedman, A.J. Nanotechnology as a therapeutic tool to combat microbial resistance. Adv. Drug Del. Rev. 2013, 65, 1803–1815. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Haiqi, G.; Li, K.; Xiang, J.; Lan, T.; Zhang, Z. Fabrication of silver nanoparticle sponge leather with durable antibacterial property. J. Coll. Interface Sci. 2018, 514, 338–348. [Google Scholar] [CrossRef] [PubMed]
- Hutchison, J.E. Greener nanoscience: A proactive approach to advancing applications and reducing implications of nanotechnology. ACS Nano 2008, 2, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Makarov, V.V.; Love, A.J.; Sinitsyna, O.V.; Makarova, S.S.; Yaminsky, I.V.; Taliansky, M.E.; Kalinina, N.O. “Green” nanotechnologies: Synthesis of metal nanoparticles using plants. Acta Nat. 2014, 6, 35–44. [Google Scholar] [CrossRef] [Green Version]
- De Matteis, V.; Cascione, M.; Toma, C.C.; Leporatti, S. Silver nanoparticles: Synthetic routes, in vitro toxicity and theranostic applications for cancer disease. Nanomaterials 2018, 8, 319. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.-Y.; Kim, M.; Shinde, S.; Saratale, R.G.; Sung, J.-S.; Ghodake, G. Temperature dependent synthesis of tryptophan-functionalized gold nanoparticles and their application in imaging human neuronal cells. ACS Sustain. Chem. Eng. 2017, 5, 7678–7689. [Google Scholar] [CrossRef]
- Raveendran, P.; Fu, J.; Wallen, S.L. Completely “green” synthesis and stabilization of metal nanoparticles. J. Am. Chem. Soc. 2003, 125, 13940–13941. [Google Scholar] [CrossRef]
- Arboleda, D.M.; Santillán, J.M.J.; Arce, V.B.; Fernández van Raap, M.B.; Muraca, D.; Fernández, M.A.; Torres Sanchez, R.M.; Schinca, D.C.; Scaffardi, L.B. A simple and “green” technique to synthesize long-term stability colloidal Ag nanoparticles: Fs laser ablation in a biocompatible aqueous medium. Mater. Charact. 2018, 140, 320–332. [Google Scholar] [CrossRef]
- Dahl, J.A.; Maddux, B.L.S.; Hutchison, J.E. Toward greener nanosynthesis. Chem. Rev. 2007, 107, 2228–2269. [Google Scholar] [CrossRef] [Green Version]
- Mickymaray, S. One-step synthesis of silver nanoparticles using saudi arabian desert seasonal plant Sisymbrium irio and antibacterial activity against multidrug-resistant bacterial strains. Biomolecules 2019, 9, 662. [Google Scholar] [CrossRef] [Green Version]
- Ghodake, G.S.; Deshpande, N.G.; Lee, Y.P.; Jin, E.S. Pear fruit extract-assisted room-temperature biosynthesis of gold nanoplates. Coll. Surfaces B Biointerfaces 2010, 75, 584–589. [Google Scholar] [CrossRef] [PubMed]
- Das, R.K.; Pachapur, V.L.; Lonappan, L.; Naghdi, M.; Pulicharla, R.; Maiti, S.; Cledon, M.; Dalila, L.M.A.; Sarma, S.J.; Brar, S.K. Biological synthesis of metallic nanoparticles: Plants, animals and microbial aspects. Nanotechnol. Environ. Eng. 2017, 2, 18. [Google Scholar] [CrossRef] [Green Version]
- Cardoso, V.S.; Quelemes, P.V.; Amorin, A.; Primo, F.L.; Gobo, G.G.; Tedesco, A.C.; Mafud, A.C.; Mascarenhas, Y.P.; Corrêa, J.R.; Kuckelhaus, S.A.S.; et al. Collagen-based silver nanoparticles for biological applications: Synthesis and characterization. J. Nanobiotechnol. 2014, 12, 36. [Google Scholar] [CrossRef] [PubMed]
- Ghodake, G.; Lim, S.-R.; Lee, D.S. Casein hydrolytic peptides mediated green synthesis of antibacterial silver nanoparticles. Coll. Surfaces B Biointerfaces 2013, 108, 147–151. [Google Scholar] [CrossRef]
- Kim, M.; Jee, S.-C.; Shinde, S.K.; Mistry, B.M.; Saratale, R.G.; Saratale, G.D.; Ghodake, G.S.; Kim, D.-Y.; Sung, J.-S.; Kadam, A.A. Green-synthesis of anisotropic peptone-silver nanoparticles and its potential application as anti-bacterial agent. Polymers 2019, 11, 271. [Google Scholar] [CrossRef] [Green Version]
- Dahoumane, S.A.; Jeffryes, C.; Mechouet, M.; Agathos, S.N. Biosynthesis of inorganic nanoparticles: A fresh look at the control of shape, size and composition. Bioengineering 2017, 4, 14. [Google Scholar] [CrossRef] [Green Version]
- Shah, M.; Fawcett, D.; Sharma, S.; Tripathy, S.K.; Poinern, G.E.J. Green synthesis of metallic nanoparticles via biological entities. Materials 2015, 8, 7278–7308. [Google Scholar] [CrossRef] [Green Version]
- Gahlawat, G.; Choudhury, A.R. A review on the biosynthesis of metal and metal salt nanoparticles by microbes. RSC Adv. 2019, 9, 12944–12967. [Google Scholar] [CrossRef] [Green Version]
- Hebbalalu, D.; Lalley, J.; Nadagouda, M.N.; Varma, R.S. Greener techniques for the synthesis of silver nanoparticles using plant extracts, enzymes, bacteria, biodegradable polymers and microwaves. ACS Sustain. Chem. Eng. 2013, 1, 703–712. [Google Scholar] [CrossRef]
- Ahmad, A.; Senapati, S.; Khan, M.I.; Kumar, R.; Sastry, M. Extracellular biosynthesis of monodisperse gold nanoparticles by a novel extremophilic actinomycete, Thermomonospora sp. Langmuir 2003, 19, 3550–3553. [Google Scholar] [CrossRef]
- Bhatnagar, S.; Kobori, T.; Ganesh, D.; Ogawa, K.; Aoyagi, H. Biosynthesis of Silver nanoparticles mediated by extracellular pigment from Talaromyces purpurogenus and their biomedical applications. Nanomaterials 2019, 9, 1042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albanese, A.; Walkey, C.D.; Olsen, J.B.; Guo, H.; Emili, A.; Chan, W.C.W. Secreted biomolecules alter the biological identity and cellular interactions of nanoparticles. ACS Nano 2014, 8, 5515–5526. [Google Scholar] [CrossRef] [PubMed]
- Heck, C.; Kanehira, Y.; Kneipp, J.; Bald, I. Amorphous carbon generation as a photocatalytic reaction on dna-assembled gold and silver nanostructures. Molecules 2019, 24, 2324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jarboe, J.K.; Mabrouk, A.F. Free amino acids, sugars and organic acids in aqueous beef extract. J. Agr. Food Chem. 1974, 22, 787–791. [Google Scholar] [CrossRef]
- Moyes, R.B.; Reynolds, J.; Breakwell, D.P. Differential staining of bacteria: Gram stain. In Current Protocols in Microbiology; John Wiley and Sons, Inc.: Hoboken, NJ, USA, 2005. [Google Scholar] [CrossRef]
- Rafique, M.; Sadaf, I.; Tahir, M.B.; Rafique, M.S.; Nabi, G.; Iqbal, T.; Sughra, K. Novel and facile synthesis of silver nanoparticles using Albizia procera leaf extract for dye degradation and antibacterial applications. Mater. Sci. Eng. C 2019, 99, 1313–1324. [Google Scholar] [CrossRef]
- Sosa, I.O.; Noguez, C.; Barrera, R.G. Optical properties of metal nanoparticles with arbitrary shapes. J. Phys. Chem. B 2003, 107, 6269–6275. [Google Scholar] [CrossRef] [Green Version]
- Roopan, S.M.; Rohit; Madhumitha, G.; Rahuman, A.; Kamaraj, C.; Bharathi, A.; Surendra, T.V. Low-cost and eco-friendly phyto-synthesis of silver nanoparticles using Cocos nucifera coir extract and its larvicidal activity. Ind. Crops Prod. 2013, 43, 631–635. [Google Scholar] [CrossRef]
- Khalil, M.M.H.; Ismail, E.H.; El-Baghdady, K.Z.; Mohamed, D. Green synthesis of silver nanoparticles using olive leaf extract and its antibacterial activity. Arab. J. Chem. 2014, 7, 1131–1139. [Google Scholar] [CrossRef] [Green Version]
- Behravan, M.; Hossein Panahi, A.; Naghizadeh, A.; Ziaee, M.; Mahdavi, R.; Mirzapour, A. Facile green synthesis of silver nanoparticles using Berberis vulgaris leaf and root aqueous extract and its antibacterial activity. Inter. J. Biol. Macromol. 2019, 124, 148–154. [Google Scholar] [CrossRef]
- Volkov, I.L.; Smirnova, A.; Makarova, A.A.; Reveguk, Z.V.; Ramazanov, R.R.; Usachov, D.Y.; Adamchuk, V.K.; Kononov, A.I. DNA with ionic, atomic and clustered silver: An XPS study. J. Phys. Chem. B 2017, 121, 2400–2406. [Google Scholar] [CrossRef]
- Fang, J.-Y.; Chen, J.-P.; Leu, Y.-L.; Wang, H.-Y. Characterization and evaluation of silk protein hydrogels for drug delivery. Chem. Pharm. Bull. 2006, 54, 156–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watson, S.M.D.; Mohamed, H.D.A.; Horrocks, B.R.; Houlton, A. Electrically conductive magnetic nanowires using an electrochemical DNA-templating route. Nanoscale 2013, 5, 5349–5359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jyoti, K.; Baunthiyal, M.; Singh, A. Characterization of silver nanoparticles synthesized using Urtica dioica Linn leaves and their synergistic effects with antibiotics. J. Rad. Res. Appl. Sci. 2016, 9, 217–227. [Google Scholar] [CrossRef] [Green Version]
- Shukla, R.; Nune, S.K.; Chanda, N.; Katti, K.; Mekapothula, S.; Kulkarni, R.R.; Welshons, W.V.; Kannan, R.; Katti, K.V. Soybeans as a phytochemical reservoir for the production and stabilization of biocompatible gold nanoparticles. Small 2008, 4, 1425–1436. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Bulut, O.; Some, S.; Mandal, A.K.; Yilmaz, M.D. Green synthesis of silver nanoparticles: Biomolecule-nanoparticle organizations targeting antimicrobial activity. RSC Adv. 2019, 9, 2673–2702. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Jun, B.-H. Silver nanoparticles: Synthesis and application for nanomedicine. Int. J. Mol. Sci. 2019, 20, 865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, W.K.; Koo, H.C.; Kim, K.W.; Shin, S.; Kim, S.H.; Park, Y.H. Antibacterial activity and mechanism of action of the silver ion in Staphylococcus aureus and Escherichia coli. Appl. Environ. Microbiol. 2008, 74, 2171–2178. [Google Scholar] [CrossRef] [Green Version]
- Padmos, J.D.; Boudreau, R.T.M.; Weaver, D.F.; Zhang, P. Structure of tiopronin-protected silver nanoclusters in a one-dimensional assembly. J. Phys. Chem. C 2015, 119, 24627–24635. [Google Scholar] [CrossRef]
- Kędziora, A.; Speruda, M.; Krzyżewska, E.; Rybka, J.; Łukowiak, A.; Bugla-Płoskońska, G. Similarities and differences between silver ions and silver in nanoforms as antibacterial agents. Int. J. Mol. Sci. 2018, 19, 444. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Yang, J.; Xie, J.; Luo, Z.; Jiang, J.; Yang, Y.Y.; Liu, S. The potent antimicrobial properties of cell penetrating peptide-conjugated silver nanoparticles with excellent selectivity for Gram-positive bacteria over erythrocytes. Nanoscale 2013, 5, 3834–3840. [Google Scholar] [CrossRef]
- Niño-Martínez, N.; Salas Orozco, M.F.; Martínez-Castañón, G.-A.; Torres Méndez, F.; Ruiz, F. Molecular mechanisms of bacterial resistance to metal and metal oxide nanoparticles. Int. J. Mol. Sci. 2019, 20, 2808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qing, Y.a.; Cheng, L.; Li, R.; Liu, G.; Zhang, Y.; Tang, X.; Wang, J.; Liu, H.; Qin, Y. Potential antibacterial mechanism of silver nanoparticles and the optimization of orthopedic implants by advanced modification technologies. Int. J. Nanomed. 2018, 13, 3311–3327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sondi, I.; Salopek-Sondi, B. Silver nanoparticles as antimicrobial agent: A case study on E. coli as a model for Gram-negative bacteria. J. Coll. Interface Sci. 2004, 275, 177–182. [Google Scholar] [CrossRef] [PubMed]








© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghodake, G.; Kim, M.; Sung, J.-S.; Shinde, S.; Yang, J.; Hwang, K.; Kim, D.-Y. Extracellular Synthesis and Characterization of Silver Nanoparticles—Antibacterial Activity against Multidrug-Resistant Bacterial Strains. Nanomaterials 2020, 10, 360. https://doi.org/10.3390/nano10020360
Ghodake G, Kim M, Sung J-S, Shinde S, Yang J, Hwang K, Kim D-Y. Extracellular Synthesis and Characterization of Silver Nanoparticles—Antibacterial Activity against Multidrug-Resistant Bacterial Strains. Nanomaterials. 2020; 10(2):360. https://doi.org/10.3390/nano10020360
Chicago/Turabian StyleGhodake, Gajanan, Min Kim, Jung-Suk Sung, Surendra Shinde, Jiwook Yang, Kyojung Hwang, and Dae-Young Kim. 2020. "Extracellular Synthesis and Characterization of Silver Nanoparticles—Antibacterial Activity against Multidrug-Resistant Bacterial Strains" Nanomaterials 10, no. 2: 360. https://doi.org/10.3390/nano10020360
APA StyleGhodake, G., Kim, M., Sung, J. -S., Shinde, S., Yang, J., Hwang, K., & Kim, D. -Y. (2020). Extracellular Synthesis and Characterization of Silver Nanoparticles—Antibacterial Activity against Multidrug-Resistant Bacterial Strains. Nanomaterials, 10(2), 360. https://doi.org/10.3390/nano10020360