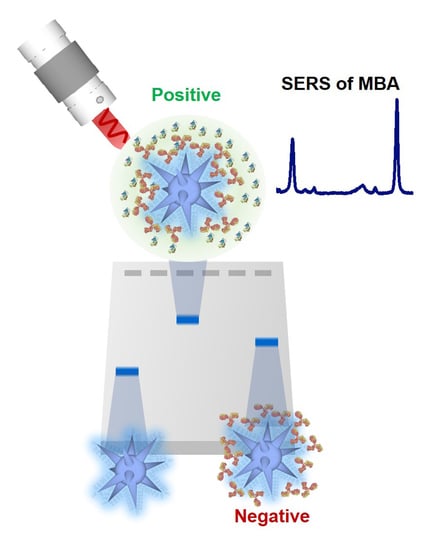
Design and Simple Assembly of Gold Nanostar Bioconjugates for Surface-Enhanced Raman Spectroscopy Immunoassays
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Materials
2.2. Gold Nanoparticles Synthesis and Functionalization
2.3. Bioconjugates Assembly
2.4. HRP Enzymatic Assay
2.5. Ultraviolet-Visible Spectroscopy
2.6. Light Scattering Measurements
2.7. Scanning Electron Microscopy and X-ray Powder Diffraction
2.8. Agarose Gel Electrophoresis
2.9. Adsorption Isotherm Fitting to AGE Data
2.10. Raman and SERS Measurements
3. Results and Discussion
3.1. Characterization of the Synthesized Gold Nanoparticles
3.2. Functionalization with the Raman Reporter
3.3. Assembly of Antibody Conjugates
3.4. Blocking of Antibody Conjugates
3.5. Competence of the Bioconjugates for SERS Immunoassays
3.5.1. Functionality of the Adsorbed Anti-HRP Antibodies
3.5.2. SERS Activity of the Bioconjugates
3.6. Proof-of-Concept for Using the Bioconjugates in the Immunoassay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Short, K.W.; Carpenter, S.; Freyer, J.P.; Mourant, J.R. Raman spectroscopy detects biochemical changes due to proliferation in mammalian cell cultures. Biophys. J. 2005, 88, 4274–4288. [Google Scholar] [CrossRef] [PubMed]
- Behl, I.; Calado, G.; Ibrahim, O.; Malkin, A.; Flint, S.; Byrne, H.J.; Lyng, F.M. Development of methodology for Raman microspectroscopic analysis of oral exfoliated cells. Anal. Methods 2017, 9, 1–22. [Google Scholar] [CrossRef]
- Farhane, Z.; Bonnier, F.; Maher, M.A.; Bryant, J.; Casey, A.; Byrne, H.J. Differentiating responses of lung cancer cell lines to Doxorubicin exposure: In vitro Raman micro spectroscopy, oxidative stress and bcl-2 protein expression. J. Biophotonics 2016, 165, 151–165. [Google Scholar] [CrossRef]
- Schlücker, S. Surface-enhanced Raman spectroscopy: Concepts and chemical applications. Angew. Chem. Int. Ed. Engl. 2014, 53, 4756–4795. [Google Scholar] [CrossRef] [PubMed]
- Howes, P.D.; Rana, S.; Stevens, M.M. Plasmonic nanomaterials for biodiagnostics. Chem. Soc. Rev. 2014, 43, 3835–3853. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zong, S.; Wu, L.; Zhu, D.; Cui, Y. SERS-Activated Platforms for Immunoassay: Probes, Encoding Methods, and Applications. Chem. Rev. 2017, 117, 7910–7963. [Google Scholar] [CrossRef] [PubMed]
- Nolan, J.P.; Duggan, E.; Condello, D. Optimization of SERS tag intensity, binding footprint, and emittance. Bioconjug. Chem. 2014, 25, 1233–1242. [Google Scholar] [CrossRef]
- Chinen, A.B.; Guan, C.M.; Ferrer, J.R.; Barnaby, S.N.; Merkel, T.J.; Mirkin, C.A. Nanoparticle Probes for the Detection of Cancer Biomarkers, Cells, and Tissues by Fluorescence. Chem. Rev. 2015, 115, 10530–10574. [Google Scholar] [CrossRef] [Green Version]
- Nolan, J.P.; Duggan, E.; Liu, E.; Condello, D.; Dave, I.; Stoner, S.A. Single cell analysis using surface enhanced Raman scattering (SERS) tags. Methods 2013, 57, 272–279. [Google Scholar] [CrossRef]
- Jett, J.H. Raman spectroscopy comes to flow cytometry. Cytom. Part A 2008, 73, 109–110. [Google Scholar] [CrossRef]
- Schütz, M.; Schlücker, S. Towards quantitative multi-color nanodiagnostics: Spectral multiplexing with six silica-encapsulated SERS labels. J. Raman Spectrosc. 2016, 47, 1012–1016. [Google Scholar] [CrossRef]
- Franco, R.; Pereira, E. Gold Nanoparticles and Proteins, Interaction. In Encyclopedia of Metalloproteins; Springer: New York, NY, USA, 2013; Volume 111, pp. 908–915. [Google Scholar]
- Shang, L.; Wang, Y.; Jiang, J.; Dong, S. pH-Dependent Protein Conformational Changes in Albumin: Gold Nanoparticle Bioconjugates: A Spectroscopic Study. Langmuir 2007, 23, 2714–2721. [Google Scholar] [CrossRef] [PubMed]
- Boulos, S.P.; Davis, T.A.; Yang, J.A.; Lohse, S.E.; Alkilany, A.M.; Holland, L.A.; Murphy, C.J. Nanoparticle–Protein Interactions: A Thermodynamic and Kinetic Study of the Adsorption of Bovine Serum Albumin to Gold Nanoparticle Surfaces. Langmuir 2013, 29, 14984–14996. [Google Scholar] [CrossRef] [PubMed]
- Cavadas, M.A.S.; Monopoli, M.P.; Cunha, C.S.E.; Prudêncio, M.; Pereira, E.; Lynch, I.; Dawson, K.A.; Franco, R. Unravelling Malaria Antigen Binding to Antibody-Gold Nanoparticle Conjugates. Part. Part. Syst. Charact. 2016, 33, 906–915. [Google Scholar] [CrossRef]
- Peixoto de Almeida, M.; Quaresma, P.; Sousa, S.; Couto, C.; Gomes, I.; Krippahl, L.; Franco, R.; Pereira, E. Measurement of adsorption constants of laccase on gold nanoparticles to evaluate the enhancement in enzyme activity of adsorbed laccase. Phys. Chem. Chem. Phys. 2018, 20, 16761–16769. [Google Scholar] [CrossRef] [PubMed]
- Käkinen, A.; Ding, F.; Chen, P.; Mortimer, M.; Kahru, A.; Ke, P.C. Interaction of firefly luciferase and silver nanoparticles and its impact on enzyme activity. Nanotechnology 2013, 24, 345101. [Google Scholar] [CrossRef]
- Ashrafpour, S.; Tohidi Moghadam, T. Interaction of silver nanoparticles with Lysozyme: Functional and structural investigations. Surf. Interfaces 2018, 10, 216–221. [Google Scholar] [CrossRef]
- Gomes, I.; Santos, N.C.; Oliveira, L.M.A.; Quintas, A.; Eaton, P.; Pereira, E.; Franco, R. Probing Surface Properties of Cytochrome c at Au Bionanoconjugates. J. Phys. Chem. C 2008, 112, 16340–16347. [Google Scholar] [CrossRef] [Green Version]
- Reymond-Laruinaz, S.; Saviot, L.; Potin, V.; de Lucas, M.D.C.M. Protein–nanoparticle interaction in bioconjugated silver nanoparticles: A transmission electron microscopy and surface enhanced Raman spectroscopy study. Appl. Surf. Sci. 2016, 389, 17–24. [Google Scholar] [CrossRef]
- Cortez, J.; Vorobieva, E.; Gralheira, D.; Osório, I.; Soares, L.; Vale, N.; Pereira, E.; Gomes, P.; Franco, R. Bionanoconjugates of tyrosinase and peptide-derivatised gold nanoparticles for biosensing of phenolic compounds. J. Nanoparticle Res. 2011, 13, 1101–1113. [Google Scholar] [CrossRef]
- Giannini, V.; Rodríguez-Oliveros, R.; Sánchez-Gil, J.A. Surface Plasmon Resonances of Metallic Nanostars/Nanoflowers for Surface-Enhanced Raman Scattering. Plasmonics 2010, 5, 99–104. [Google Scholar] [CrossRef] [Green Version]
- Ma, W.Y.; Yang, H.; Hilton, J.P.; Lin, Q.; Liu, J.Y.; Huang, L.X.; Yao, J. A numerical investigation of the effect of vertex geometry on localized surface plasmon resonance of nanostructures. Opt. Express 2010, 18, 843–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nehl, C.L.; Liao, H.; Hafner, J.H. Optical Properties of Star-Shaped Gold Nanoparticles. Nano Lett. 2006, 6, 683–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, M.J.; Quaresma, P.; De Almeida, M.P.; Araújo, A.; Pereira, E.; Fortunato, E.; Martins, R.; Franco, R.; Águas, H. Office paper decorated with silver nanostars-an alternative cost effective platform for trace analyte detection by SERS. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Depciuch, J.; Stec, M.; Maximenko, A.; Pawlyta, M.; Baran, J.; Parlinska-Wojtan, M. Control of Arms of Au Stars Size and its Dependent Cytotoxicity and Photosensitizer Effects in Photothermal Anticancer Therapy. Int. J. Mol. Sci. 2019, 20, 1–14. [Google Scholar] [CrossRef]
- Puertas, S.; Batalla, P.; Moros, M.; Polo, E.; del Pino, P.; Guisán, J.M.; Grazú, V.; de la Fuente, J.M. Taking Advantage of Unspecific Interactions to Produce Highly Active Magnetic Nanoparticle−Antibody Conjugates. ACS Nano 2011, 5, 4521–4528. [Google Scholar] [CrossRef]
- Smith, P.K.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.D.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]
- Ojea-Jiménez, I.; Bastús, N.G.; Puntes, V. Influence of the sequence of the reagents addition in the citrate-mediated synthesis of gold nanoparticles. J. Phys. Chem. C 2011, 115, 15752–15757. [Google Scholar] [CrossRef]
- Yuan, H.; Khoury, C.G.; Hwang, H.; Wilson, C.M.; Grant, G.A.; Vo-Dinh, T. Gold nanostars: Surfactant-free synthesis, 3D modelling, and two-photon photoluminescence imaging. Nanotechnology 2012, 23, 075102. [Google Scholar] [CrossRef]
- Haiss, W.; Thanh, N.T.K.; Aveyard, J.; Fernig, D.G. Determination of Size and Concentration of Gold Nanoparticles from UV—Vis Spectra. Anal. Chem. 2007, 79, 4215–4221. [Google Scholar] [CrossRef]
- De Puig, H.; Tam, J.O.; Yen, C.-W.; Gehrke, L.; Hamad-Schifferli, K. Extinction Coefficient of Gold Nanostars. J. Phys. Chem. C 2015, 119, 17408–17415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sigma-Aldrich Enzymatic Assay of Peroxidase (EC 1.11.1.7) 2,2′-Azino-bis(3-Ethylbenzthiazoline-6-Sulfonic Acid) as a Substrate. Available online: https://www.sigmaaldrich.com/technical-documents/protocols/biology/enzymatic-assay-of-peroxidase-abts-as-substrate.html (accessed on 29 July 2019).
- Kim, J.-Y.; Kim, H.-B.; Jang, D.-J. Electrophoretic separation of gold nanoparticles according to bifunctional molecules-induced charge and size. Electrophoresis 2013, 34, 911–916. [Google Scholar] [CrossRef] [PubMed]
- Hanauer, M.; Pierrat, S.; Zins, I.; Lotz, A.; Sönnichsen, C. Separation of Nanoparticles by Gel Electrophoresis According to Size and Shape. Nano Lett. 2007, 7, 2881–2885. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, M.S.; Luque-Alled, J.M.; Gomez, T.; Castillo, J.R. Evaluation of agarose gel electrophoresis for characterization of silver nanoparticles in industrial products. Electrophoresis 2016, 37, 1376–1383. [Google Scholar] [CrossRef]
- Piantanida, L.; Naumenko, D.; Lazzarino, M. Highly efficient gold nanoparticle dimer formation via DNA hybridization. RSC Adv. 2014, 4, 15281–15287. [Google Scholar] [CrossRef]
- Rothe, G.M. Porosity Gradient Gels. Encycl. Sep. Sci. 2000, 13, 1315–1342. [Google Scholar]
- Xu, S.; Ji, X.; Xu, W.; Zhao, B.; Dou, X.; Bai, Y.; Ozaki, Y. Surface-enhanced Raman scattering studies on immunoassay. J. Biomed. Opt. 2005, 10, 031112-1–031112-12. [Google Scholar] [CrossRef]
- Mir-Simon, B.; Reche-Perez, I.; Guerrini, L.; Pazos-Perez, N.; Alvarez-Puebla, R.A. Universal One-Pot and Scalable Synthesis of SERS Encoded Nanoparticles. Chem. Mater. 2015, 27, 950–958. [Google Scholar] [CrossRef]
- Ahmed, W.; Stefan Kooij, E.; van Silfhout, A.; Poelsema, B. Controlling the morphology of multi-branched gold nanoparticles. Nanotechnology 2010, 21, 125605. [Google Scholar] [CrossRef]
- Garcia-Leis, A.; Torreggiani, A.; Garcia-Ramos, J.V.; Sanchez-Cortes, S. Hollow Au/Ag nanostars displaying broad plasmonic resonance and high surface-enhanced Raman sensitivity. Nanoscale 2015, 7, 13629–13637. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Leis, A.; Garcia-Ramos, J.V.; Sanchez-Cortes, S. Silver Nanostars with High SERS Performance. J. Phys. Chem. C 2013, 117, 7791–7795. [Google Scholar] [CrossRef]
- Tsoulos, T.V.; Han, L.; Weir, J.; Xin, H.L.; Fabris, L. A closer look at the physical and optical properties of gold nanostars: An experimental and computational study. Nanoscale 2017, 9, 3766–3773. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Lee, S.; Lee, J.H.; Lim, H.W.; Seong, G.H.; Lee, E.K.; Chang, S.I.; Oh, C.H.; Choo, J. Highly reproducible immunoassay of cancer markers on a gold-patterned microarray chip using surface-enhanced Raman scattering imaging. Biosens. Bioelectron. 2011, 26, 2135–2141. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, S.; Agrawal, A.; Rodríguez-Lorenzo, L.; Pastoriza-Santos, I.; Alvarez-Puebla, R.A.; Kornowski, A.; Weller, H.; Liz-Marzán, L.M. Tuning size and sensing properties in colloidal gold nanostars. Langmuir 2010, 26, 14943–14950. [Google Scholar] [CrossRef]
- Li, R.; Lv, H.; Zhang, X.; Liu, P.; Chen, L.; Cheng, J.; Zhao, B. Vibrational spectroscopy and density functional theory study of 4-mercaptobenzoic acid. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 148, 369–374. [Google Scholar] [CrossRef]
- Sánchez-Cortés, S.; García-Ramos, J.V. Anomalous Raman bands appearing in surface-enhanced Raman spectra. J. Raman Spectrosc. 1998, 29, 365–371. [Google Scholar] [CrossRef]
- Pfeiffer, C.; Rehbock, C.; Hühn, D.; Carrillo-Carrion, C.; de Aberasturi, D.J.; Merk, V.; Barcikowski, S.; Parak, W.J. Interaction of colloidal nanoparticles with their local environment: The (ionic) nanoenvironment around nanoparticles is different from bulk and determines the physico-chemical properties of the nanoparticles. J. R. Soc. Interface 2014, 11, 20130931. [Google Scholar] [CrossRef]
- Sidorov, I.A.; Prabakaran, P.; Dimitrov, D.S. Non-Covalent Conjugation of Nanoparticles to Antibodies via Electrostatic Interactions—A Computational Model. J. Comput. Theor. Nanosci. 2007, 4, 1103–1107. [Google Scholar] [CrossRef]
- Rabe, M.; Verdes, D.; Seeger, S. Understanding protein adsorption phenomena at solid surfaces. Adv. Colloid Interface Sci. 2011, 162, 87–106. [Google Scholar] [CrossRef] [Green Version]







© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliveira, M.J.; P. de Almeida, M.; Nunes, D.; Fortunato, E.; Martins, R.; Pereira, E.; J. Byrne, H.; Águas, H.; Franco, R. Design and Simple Assembly of Gold Nanostar Bioconjugates for Surface-Enhanced Raman Spectroscopy Immunoassays. Nanomaterials 2019, 9, 1561. https://doi.org/10.3390/nano9111561
Oliveira MJ, P. de Almeida M, Nunes D, Fortunato E, Martins R, Pereira E, J. Byrne H, Águas H, Franco R. Design and Simple Assembly of Gold Nanostar Bioconjugates for Surface-Enhanced Raman Spectroscopy Immunoassays. Nanomaterials. 2019; 9(11):1561. https://doi.org/10.3390/nano9111561
Chicago/Turabian StyleOliveira, Maria João, Miguel P. de Almeida, Daniela Nunes, Elvira Fortunato, Rodrigo Martins, Eulália Pereira, Hugh J. Byrne, Hugo Águas, and Ricardo Franco. 2019. "Design and Simple Assembly of Gold Nanostar Bioconjugates for Surface-Enhanced Raman Spectroscopy Immunoassays" Nanomaterials 9, no. 11: 1561. https://doi.org/10.3390/nano9111561
APA StyleOliveira, M. J., P. de Almeida, M., Nunes, D., Fortunato, E., Martins, R., Pereira, E., J. Byrne, H., Águas, H., & Franco, R. (2019). Design and Simple Assembly of Gold Nanostar Bioconjugates for Surface-Enhanced Raman Spectroscopy Immunoassays. Nanomaterials, 9(11), 1561. https://doi.org/10.3390/nano9111561